Peer Review
We believe that transparency in peer review can improve the speed, quality, and collegiality of the publication process. Peer review is also an important training opportunity for lab members in scientific thinking and writing. As such, we only review manuscripts that have been posted on preprint servers and we post our peer review comments publicly. Additional resources can be found in our Lab Compact and in the Peer Review minicourse. We support other initiatives to improve peer review (such as Review Commons and FeedbackASAP).
FusOn-pLM: A Fusion Oncoprotein-Specific Language Model via Focused Probabilistic Masking.
Vincoff S, Goel S, Kholina K, Pulugurta R, Vure P, Chatterjee P.
Reviewed by: Bajaj P, Macdonald CB.
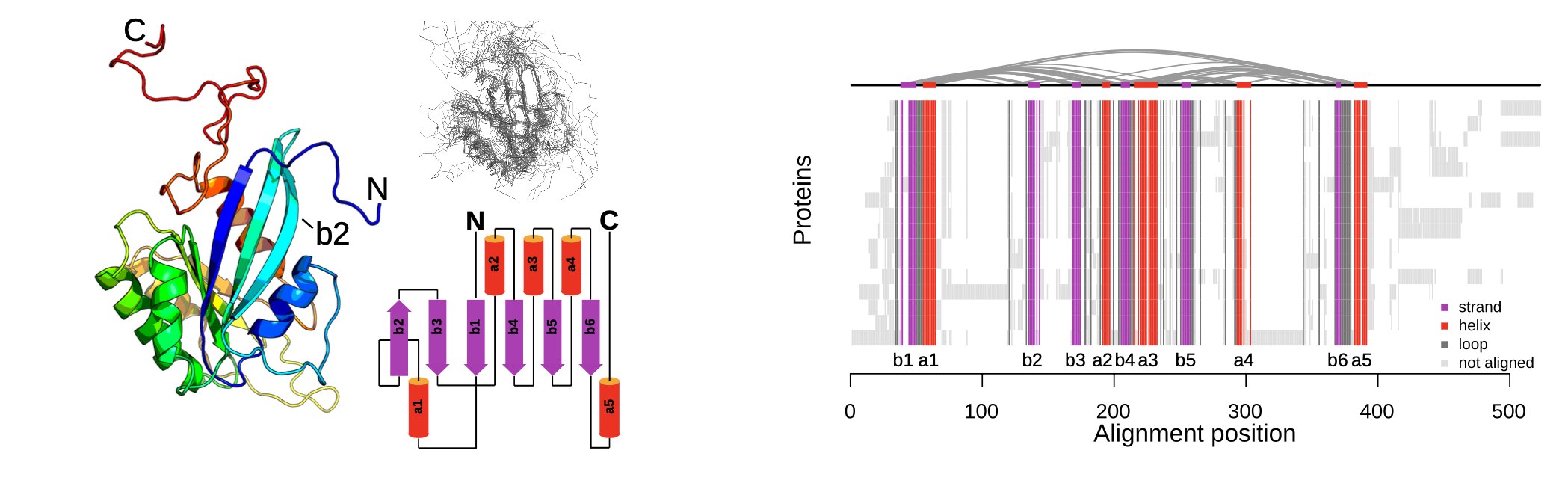
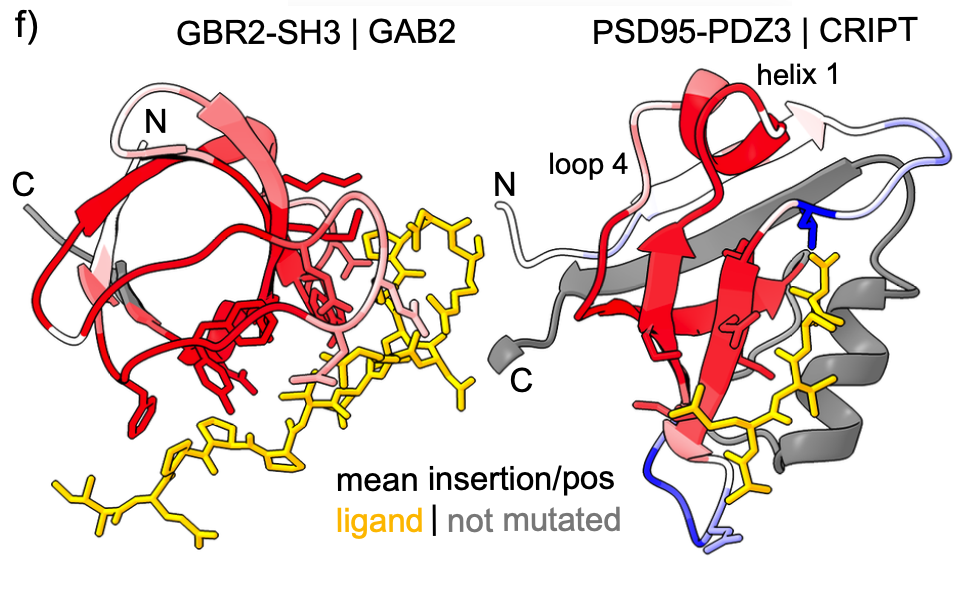
A complete map of specificity encoding for a partially fuzzy protein interaction.
Zarin T, Lehner B.
Reviewed by: Krupkin B, Le M, Coyote-Maestas W, Fraser JS.
Dissecting translation elongation dynamics through ultra-long tracking of single ribosomes.
Madern MF, Yang S, Witteveen O, Bauer M, Tanenbaum ME.
Reviewed by: Chen Y, Student 1, Student 2, Coyote-Maestas W, Fraser JS.
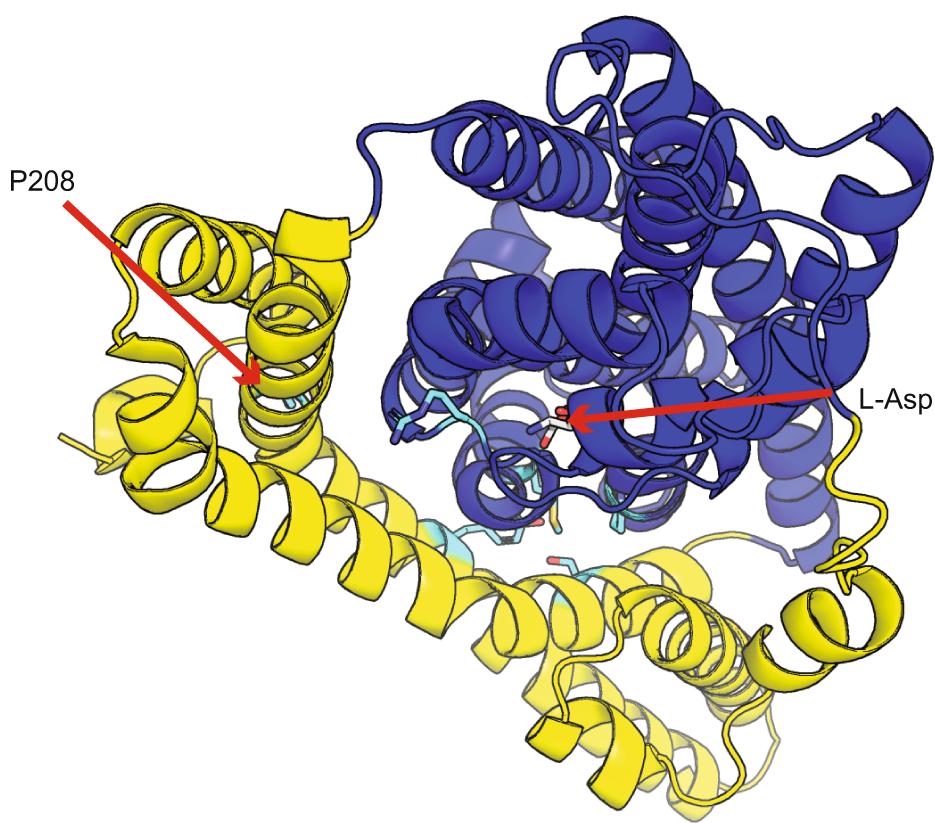
Highly multiplexed design of an allosteric transcription factor to sense novel ligands.
Nishikawa KK, Chen J, Acheson JF, Harbaugh SV, Huss P, Frenkel M, Novy N, Sieren HR, Lodewyk EC, Lee DH, Chávez JL, Fox BG, Raman S.
Reviewed by: Mullin-Bernstein Z, Chrispens K, Coyote-Maestas W, Fraser JS.
Targeting protein-ligand neosurfaces using a generalizable deep learning approach.
Marchand A, Buckley S, Schneuing A, Pacesa M, Gainza P, Elizarova E, Neeser RM, Lee PW, Reymond L, Elia M, Scheller L, Georgeon S, Schmidt J, Schwaller P, Maerkl SJ, Bronstein M, Correia BE.
Reviewed by: Chen MN, Alamo KAE, Coyote-Maestas W, Fraser JS.
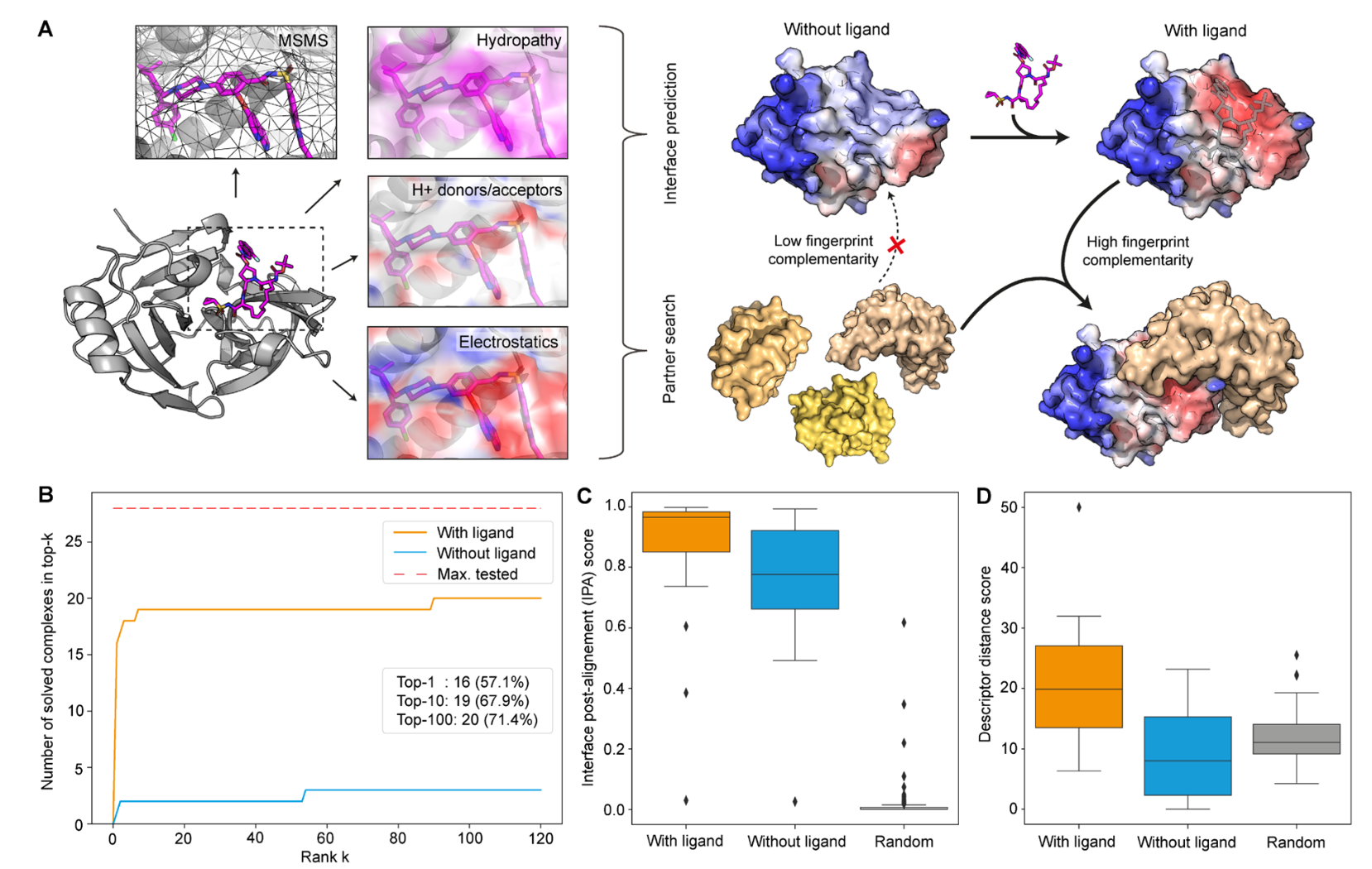
Mutational Profiling of SARS-CoV-2 PLpro in human cells reveals requirements for function, structure, and drug escape.
Wu X, Go M, Nguyen JV, Kuchel NW, Lu BG, Lowes KN, Calleja DJ, Mitchell JP, Lessene G, Komander D, Call ME.
Reviewed by: Bajaj P, Fraser JS.
Heterogeneous folding landscapes and predetermined breaking points within a protein family.
Pechmann S.
Reviewed by: Macdonald CB and San Felipe CJ.
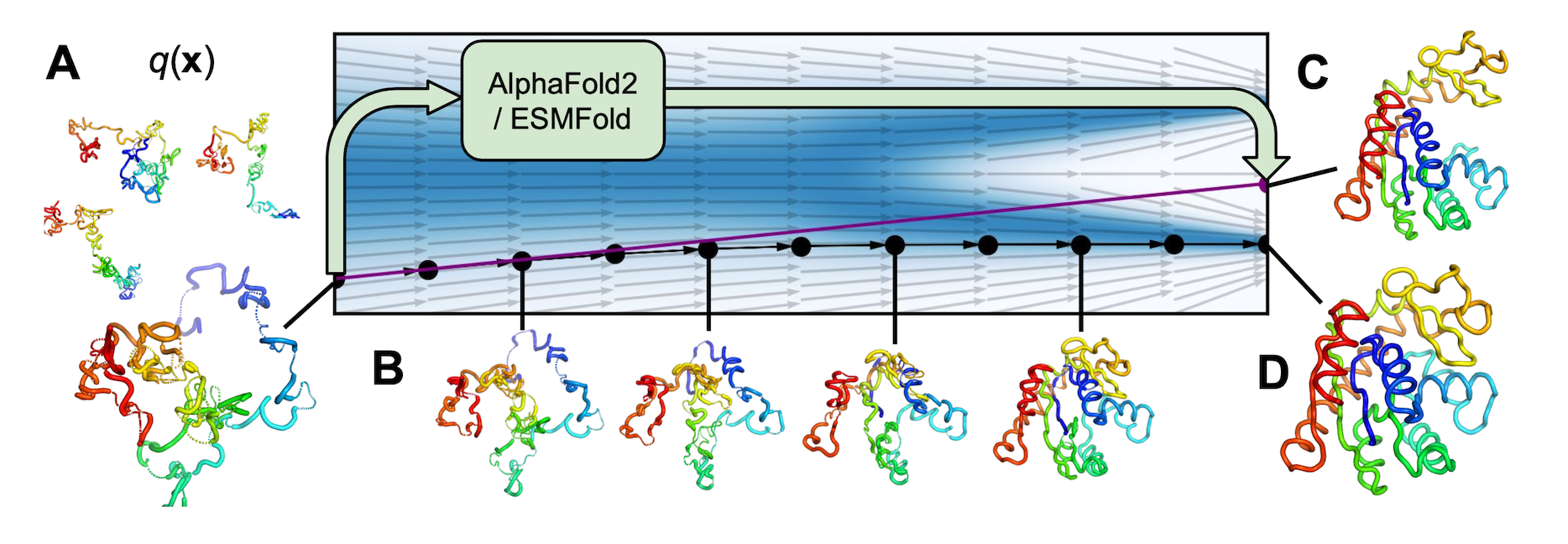
AlphaFold Meets Flow Matching for Generating Protein Ensembles.
Jing B, Berger B, Jaakkola T.
Reviewed by: Jansen F, Ravikumar A, Wankowicz SA, Fraser JS.
Prediction of Ca 2+ binding site in proteins with a fast and accurate method based on statistical mechanics and analysis of crystal structures.
Basit A, Choudhury D, Bandyopadhyay P.
Reviewed by: Wankowicz SA.
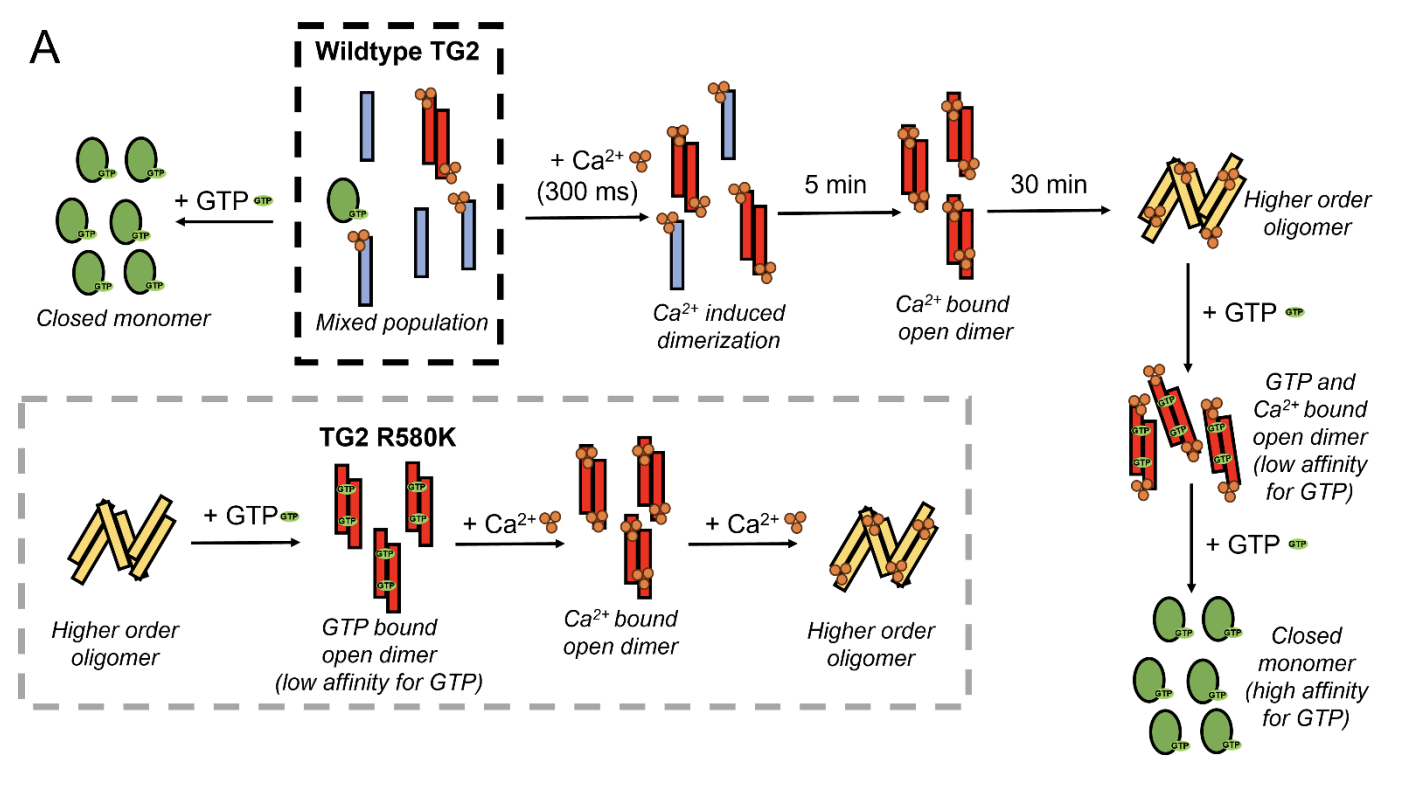
Defining the conformational states that enable transglutaminase 2 to promote cancer cell survival versus cell death.
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK.
Reviewed by: Yamamura H, Mitra R, and Fraser JS.
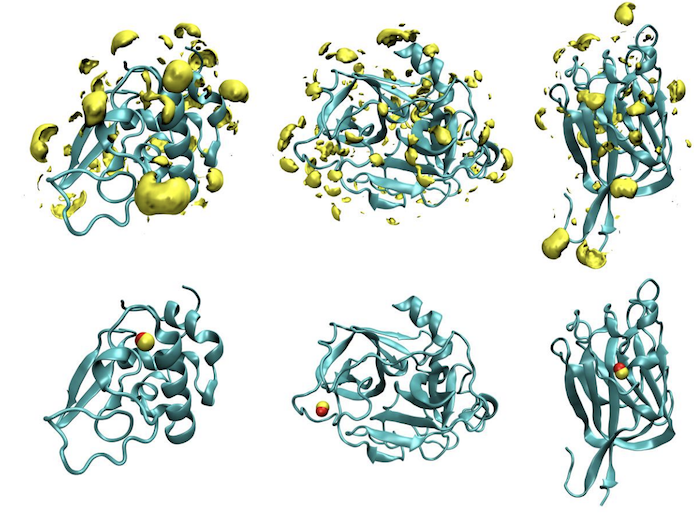
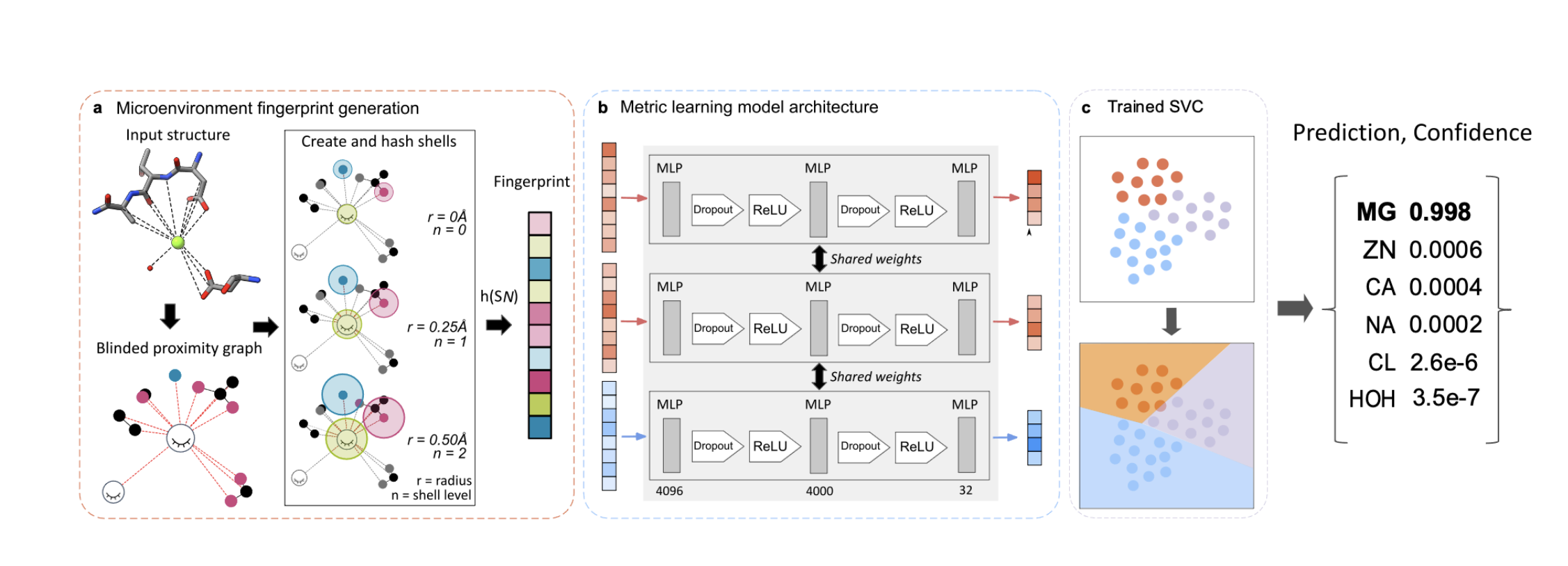
Metric Ion Classification (MIC): A deep learning tool for assigning ions and waters in cryo-EM and x-ray crystallography structures.
Shub L, Liu W, Skiniotis G, Keiser M, Robertson M.
Reviewed by: Wankowicz SA.
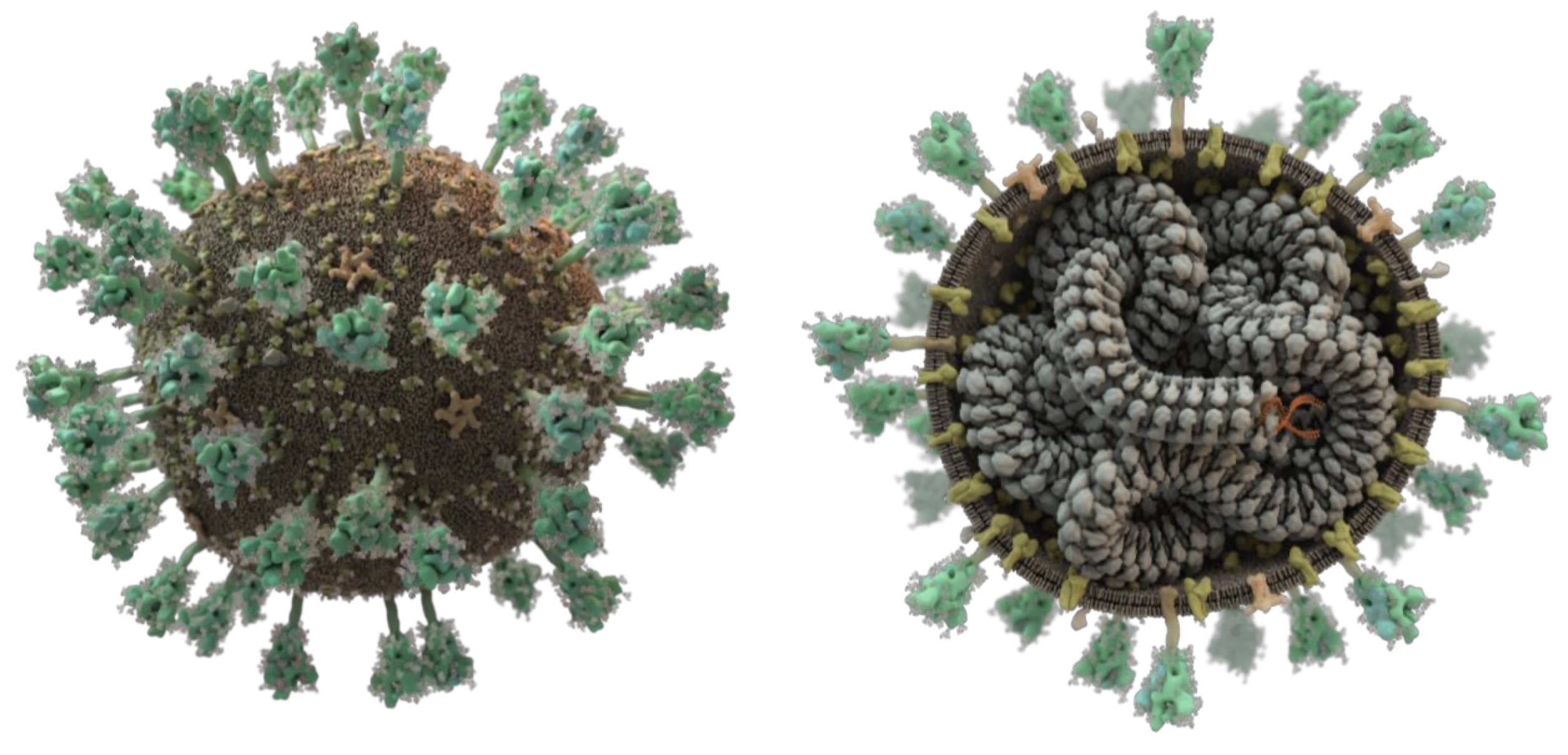
Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Ke Z, Peacock TP, Brown JC, Sheppard CM, Croll TI, Kotecha A, Goldhill DH, Barclay WS, Briggs JAG.
Reviewed by: Fraser JS, HHMI TAP participants.
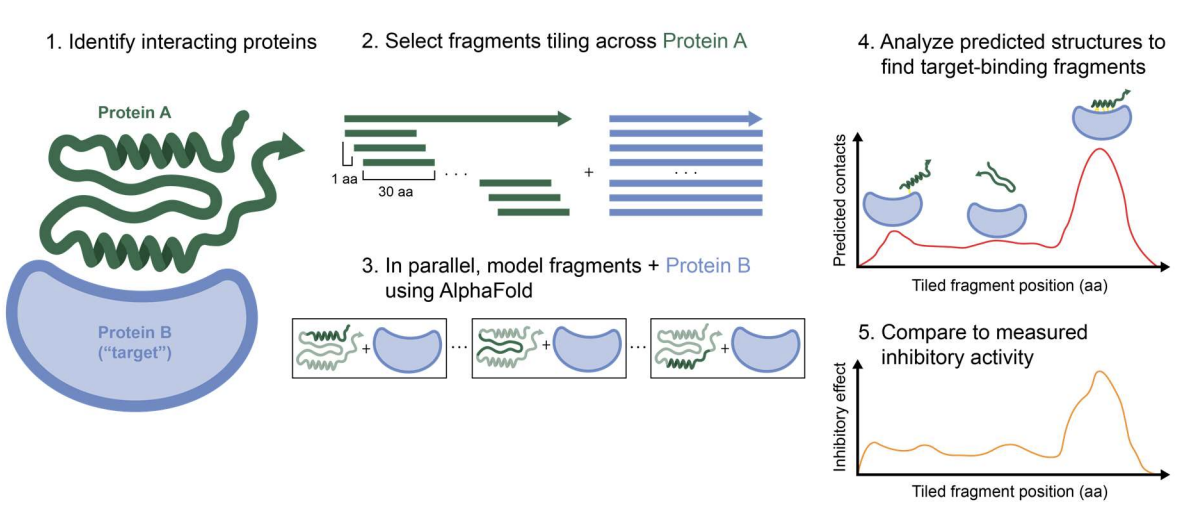
High-throughput computational discovery of inhibitory protein fragments with AlphaFold.
Savinov A, Swanson S, Keating AE, Li GW.
Reviewed by: San Felipe CJ, Bajaj P, Fraser JS.
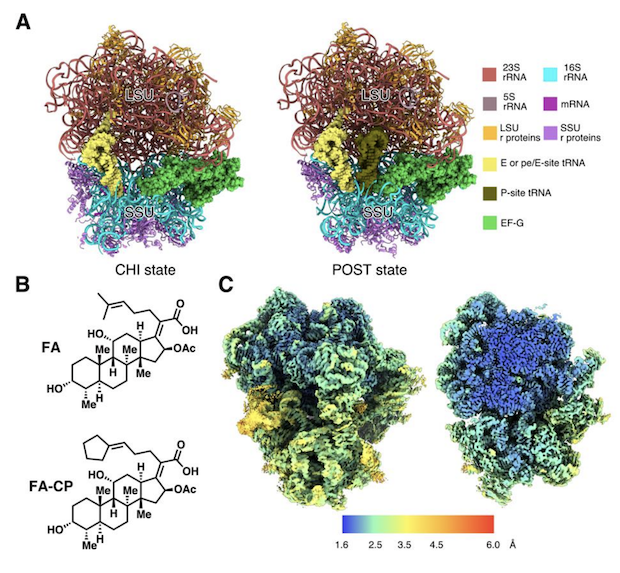
Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane.
González-López A, Larsson DSD, Koripella RK, Cain BN, Chavez MG, Hergenrother PJ, Sanyal S, Selmer M.
Reviewed by: Dandan M, Chen D, Fraser JS.
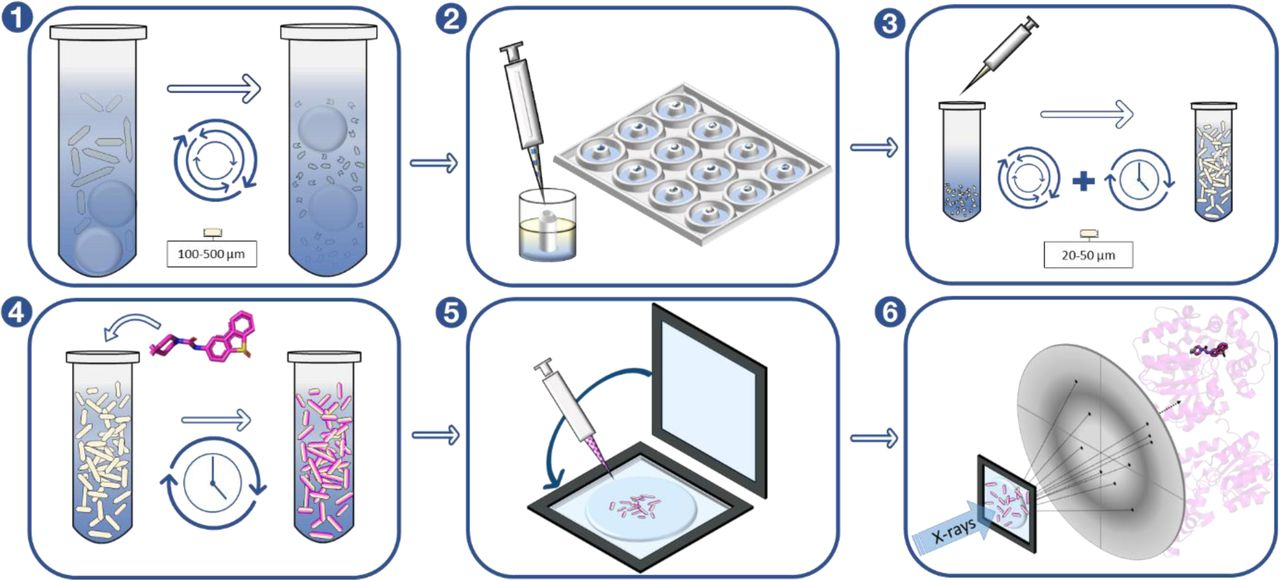
Exploring serial crystallography for drug discovery.
Dunge A, Phan C, Uwangue O, Bjelcic M, Gunnarsson J, Wehlander G, Käck H, Brändén G.
Reviewed by: Fraser JS.
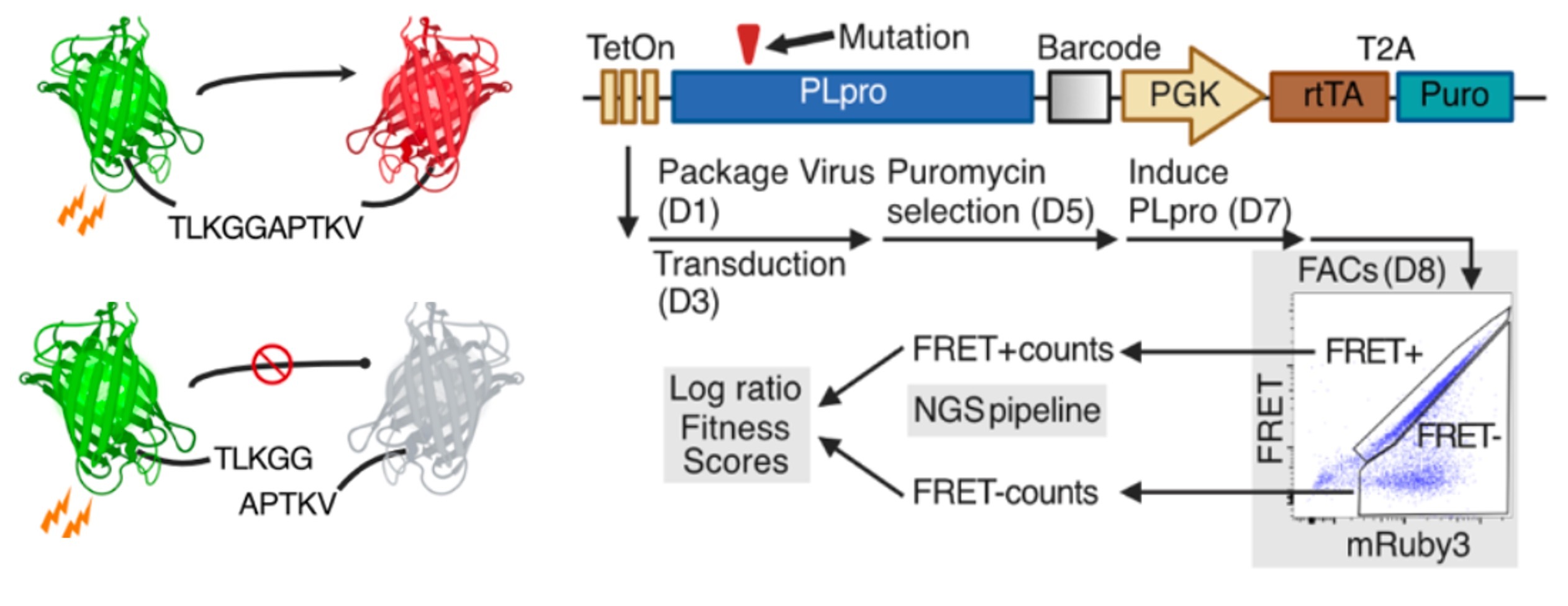
Deep indel mutagenesis reveals the impact of insertions and deletions on protein stability and function.
Topolska M, Beltran A, Lehner B.
Reviewed by: Bajaj P, Chrispens K, Fraser JS, Coyote-Maestas W.
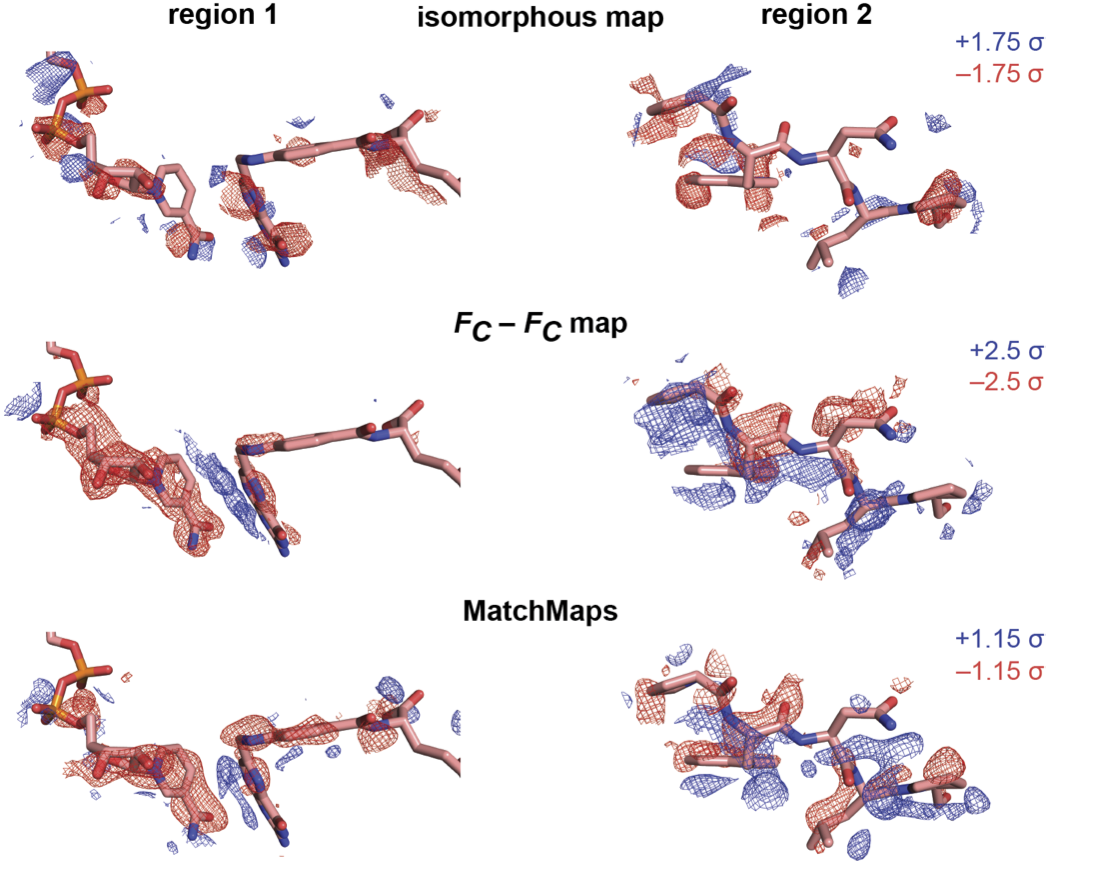
MatchMaps: Non-isomorphous difference maps for X-ray crystallography.
Brookner DE, Hekstra DR.
Reviewed by: Asthana P, Correy GJ, Fraser JS.
Approximating conformational Boltzmann distributions with AlphaFold2 predictions.
Brown BP, Stein RA, Meiler J, Mchaourab H.
Reviewed by: Flowers J, Lam A, Ravikumar A, Fraser J.
Detuning of the Ribosome Conformational Landscape Promotes Antibiotic Resistance and Collateral Sensitivity.
Mesa P, Jiménez-Fernández A, La Rosa R, Espinosa R, Johansen HK, Molin S, Montoya G.
Reviewed by: Raskar T, Dandan M, Fraser JS.
Cooperative conformational transitions and the temperature dependence of enzyme catalysis.
Walker EJ, Hamill CJ, Crean R, Connolly MS, Warrender AK, Kraakman KL, Prentice EJ, Steyn-Ross A, Steyn-Ross M, Pudney CR, van der Kamp MW, Schipper LA, Mulholland AJ, Arcus VL..
Reviewed by: Chen D, Muir D, Pinney M, Fraser JS.
Predicting Relative Populations of Protein Conformations without a Physics Engine Using AlphaFold2.
da Silva GM, Cui JY, Dalgarno DC, Lisi GP, Rubenstein BM.
Reviewed by: Ravikumar A, Lee S, Fraser Lab.
- Biorxiv Preprint v550545: 550545
- PREreview.org: 8235543
- Link: https://github.com/GMdSilva/10.5281-zenodo-8235542_response
Peer Review
Author response
Read the response
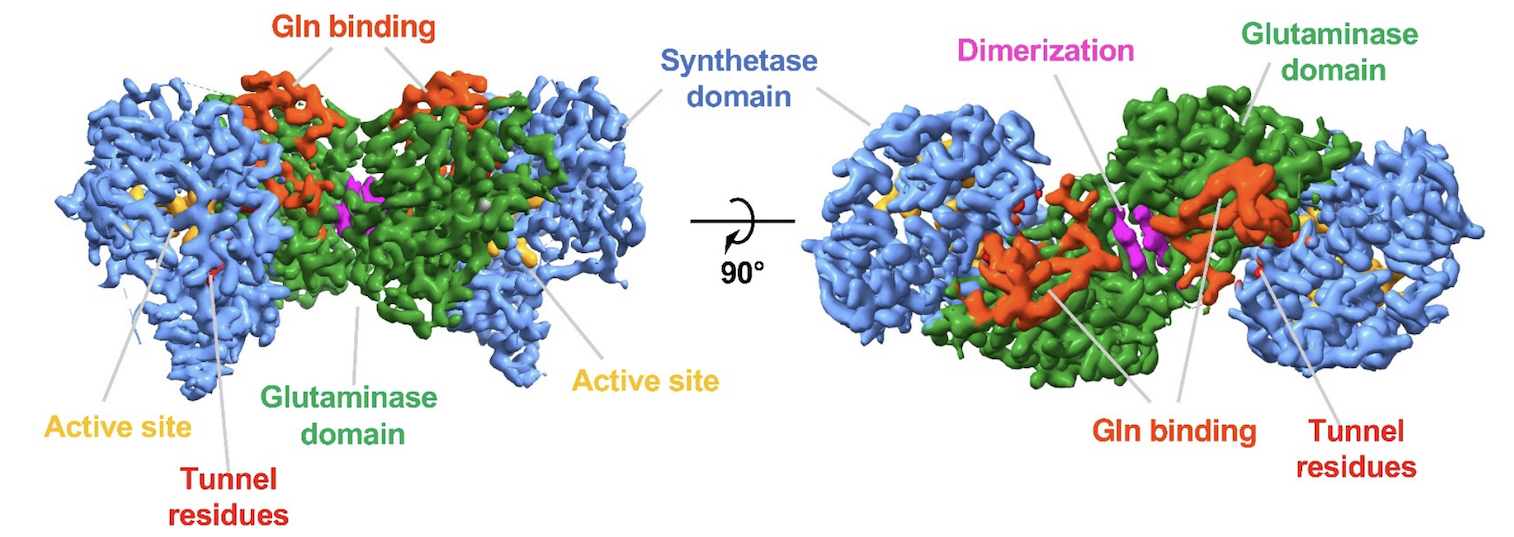
Cryo-EM and Molecular Dynamics Simulations Reveal Hidden Conformational Dynamics Controlling Ammonia Transport in Human Asparagine Synthetase.
Coricello A, Zhu W, Lupia A, Gratteri C, Vos M, Chaptal V, Alcaro S, Takagi Y, Richards N.
Reviewed by: Wankowicz S, Fraser JS.
The mineralocorticoid receptor forms higher order oligomers upon DNA binding.
Fettweis G, Johnson TA, Almeida-Prieto B, Presman DM, Hager GL, Alvarez de la Rosa D.
Reviewed by: Wankowicz S, Bergmann L, Fraser JS.
Using macromolecular electron densities to improve the enrichment of active compounds in virtual screening.
Ma W, Zhang W, Le Y, Shi X, Xu Q, Xiao Y, Dou Y, Wang X, Zhou W, Peng W, Zhang H, Huang B.
Reviewed by: San Felipe CJ, Yamamura H, Fraser J.
Computational Scoring and Experimental Evaluation of Enzymes Generated by Neural Networks.
Johnson SR, Fu X, Viknander S, Goldin C, Monaco S, Zelezniak A, Yang KK.
Reviewed by: Beazer J, Fraser JS.
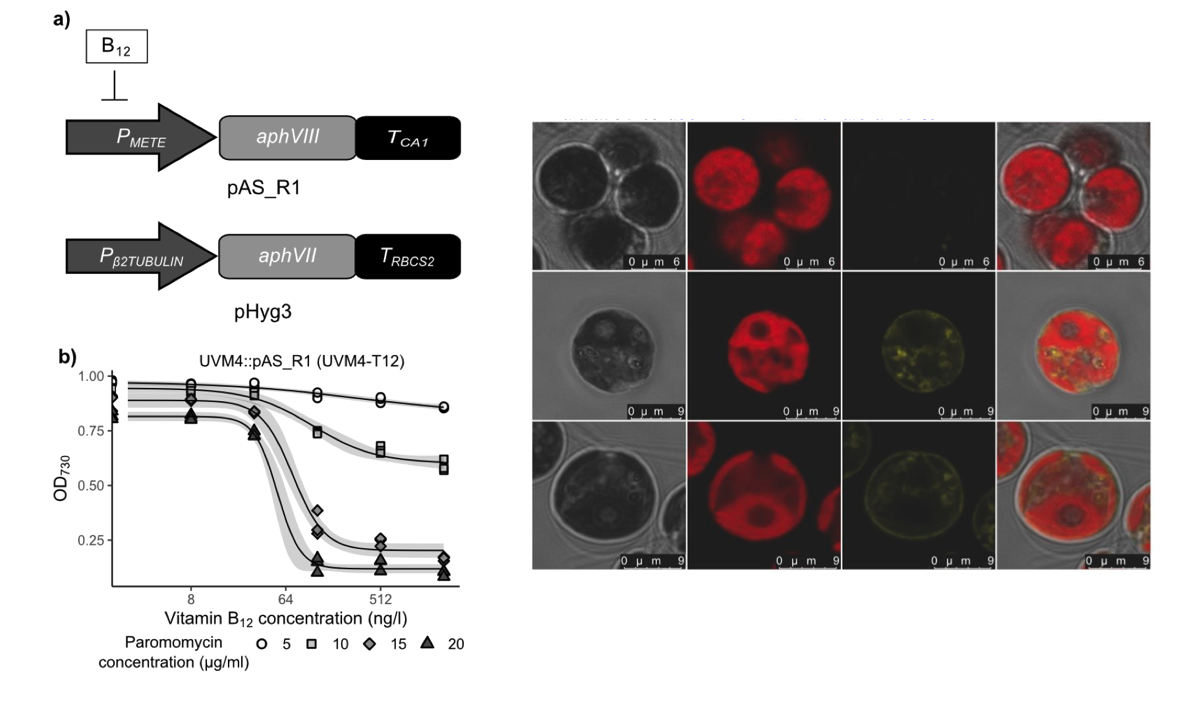
The conserved protein CBA1 is required for vitamin B12 uptake in different algal lineages.
Sayer AP, Llavero-Pasquina M, Geisler K, Holzer A, Bunbury F, Mendoza-Ochoa GI, Lawrence AD, Warren MJ, Mehrshahi P, Smith AG.
Reviewed by: Student 1, Student 2, Macdonald CB, Fraser J.
Phosphorylation of pyruvate dehydrogenase marks the inhibition of in vivo neuronal activity.
Yang D, Wang Y, Qi T, Zhang X, Shen L, Ma J, Pang Z, Lal NK, McClatchy DB, Wang K, Xie Y, Polli F, Maximov A, Augustine V, Cline HT, Yates JR III, Ye L.
Reviewed by: Liu Z, and Shin C, Macdonald CB, Fraser J.
- Biorxiv Preprint v1: 532494
- PREreview.org: doi-10.1101-2023.03.13.532494
Peer Review
- PMID: 38266644
Post-Review
Access the paper
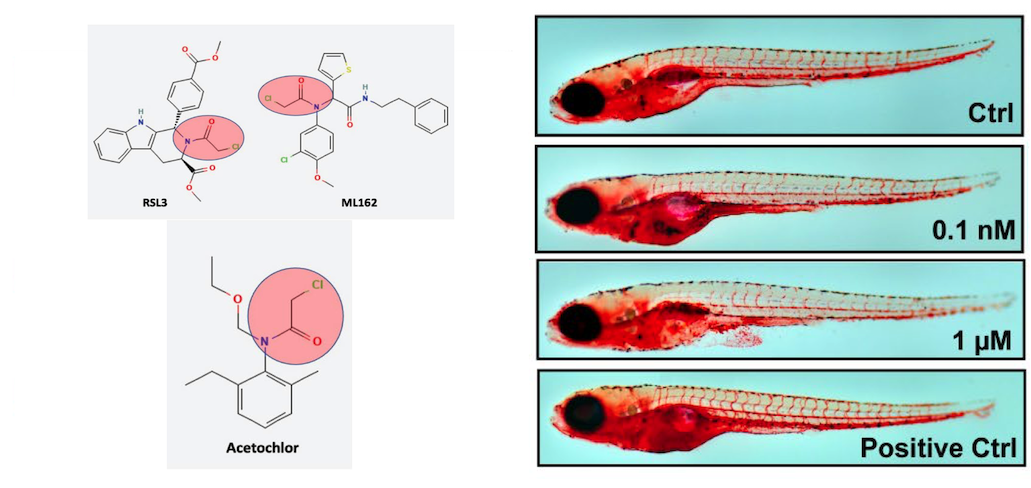
The Herbicide Acetochlor Causes Lipid Peroxidation by Inhibition of Glutathione Peroxidase 4.
Mesmar F, Muhsen M, Tourigny JP, Tennessen JM, Bondesson M.
Reviewed by: Byun D, Zheng J, Macdonald CB, Fraser J.
Decoding the Cure-all Effects of Ginseng.
Loo S, Kam A, Dutta B, Zhang X, Feng N, Sze SK, Liu CF, Wang X, Tam JP.
Reviewed by: Chavez I, Rodea D, Ravikumar A, Macdonald CB, Fraser J.
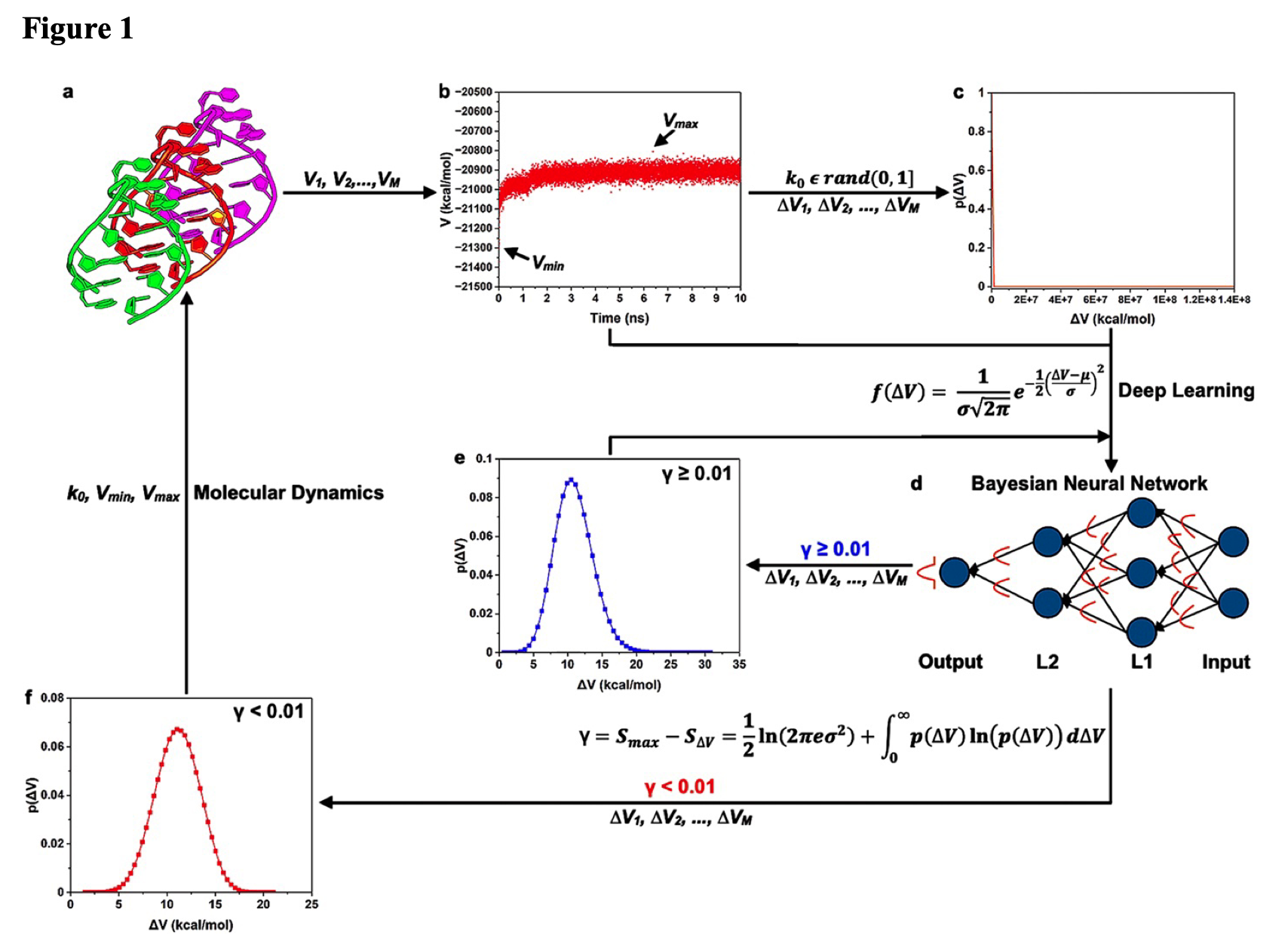
Deep Boosted Molecular Dynamics (DBMD): Accelerating molecular simulations with Gaussian boost potentials generated using probabilistic Bayesian deep neural network.
Do, H and Miao, Y.
Reviewed by: Alkislar I, Freitas N, Macdonald CB, Fraser J.
Quantification of gallium cryo-FIB milling damage in biological lamella.
Lucas BA, Grigorieff N.
Reviewed by: Raskar T, Chen D, Fraser JS.
- Biorxiv Preprint v1: 526705
- Review: 2u852i1
- Response: 2ub8gsu
Peer Review
Author response
Read the response
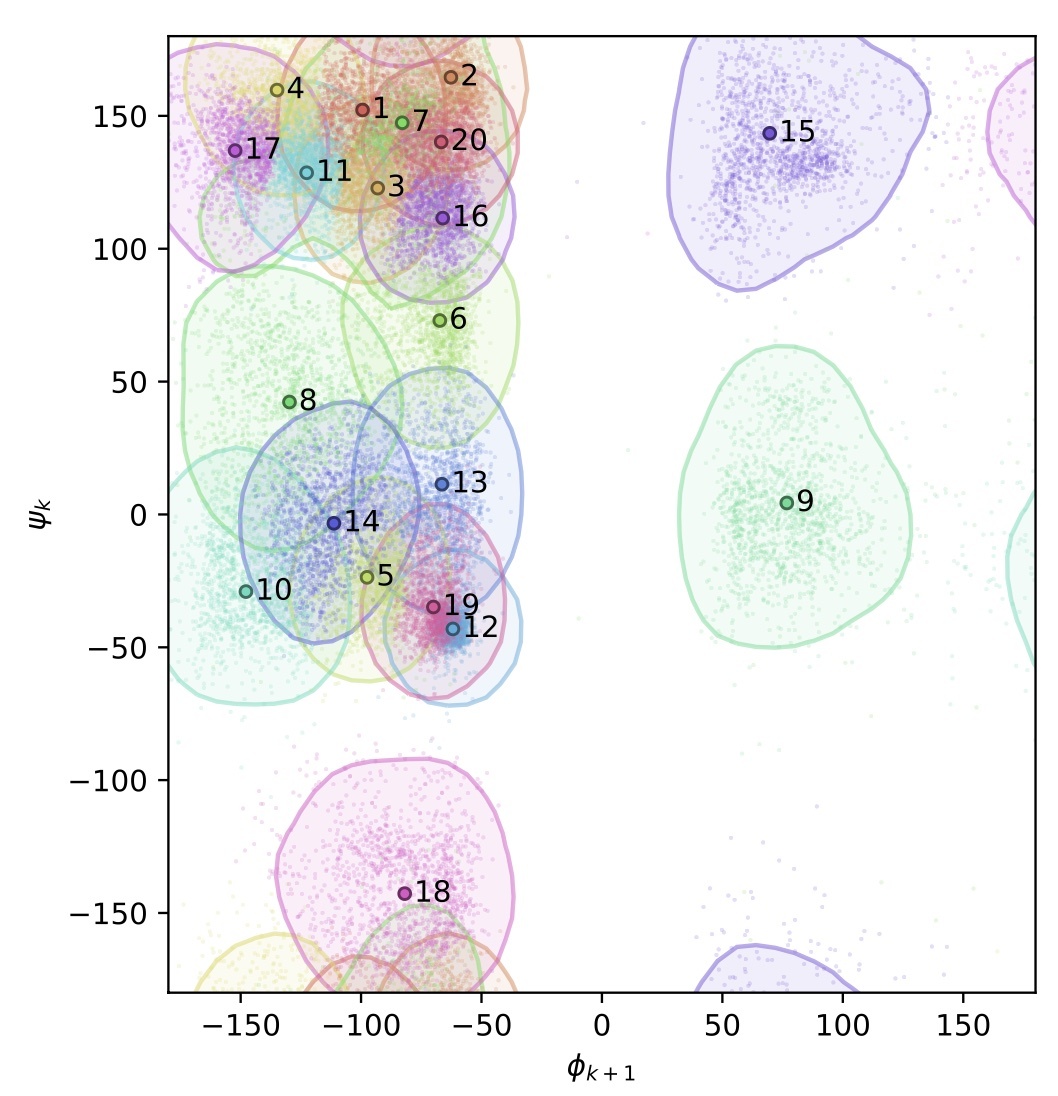
An amino domino model described by a cross peptide bond Ramachandran plot defines amino acid pairs as local structural units.
Rosenberg AA, Yehishalom N, Marx A, Bronstein A.
Reviewed by: Ravikumar A, Fraser JS.
Mutation in glutamate transporter homologue GltTk provides insights into pathologic mechanism of episodic ataxia 6.
Colucci E, Anshari ZR, Patiño-Ruiz MF, Nemchinova M, Whittaker J, Slotboom DJ, Guskov A.
Reviewed by: Macdonald CB.
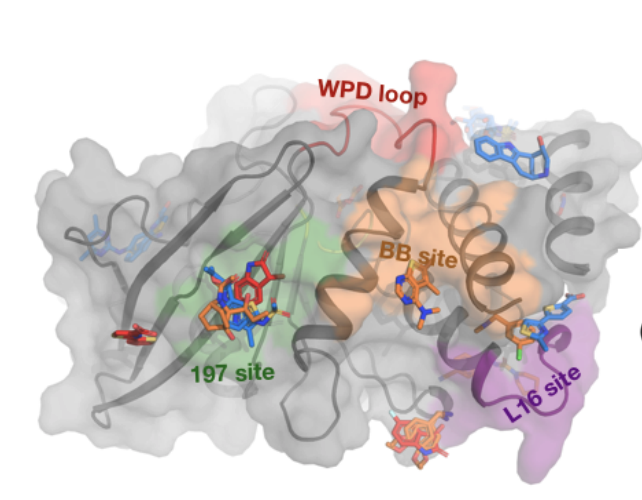
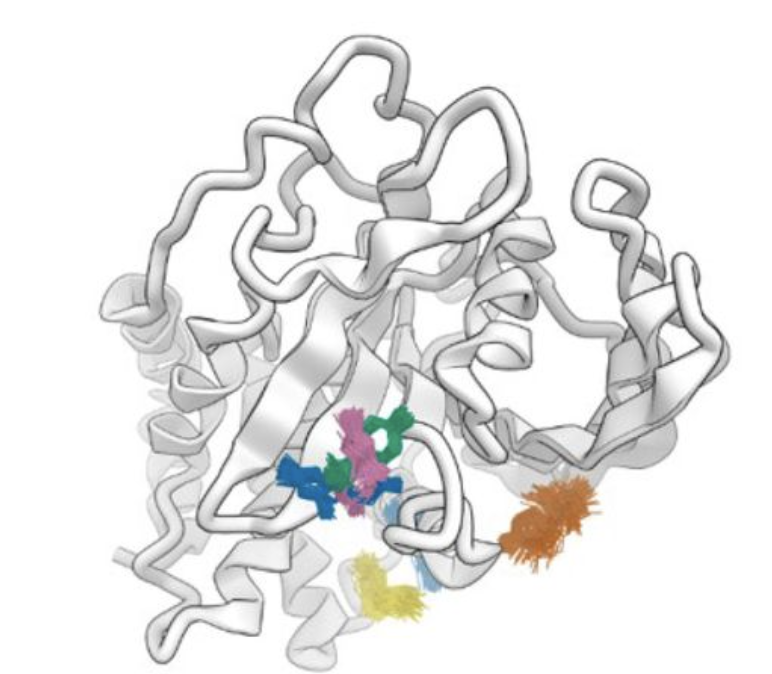
A conserved local structural motif controls the kinetics of PTP1B catalysis.
Yeh CY, Izaguirre JA, Greisman JB, Willmore L, Maragakis P, Shaw DE.
Reviewed by: San Felipe CJ, Fraser J.
- Biorxiv Preprint v1: 529746
- Review: 2tjkmcx
- PREreview.org: doi-10.1101-2023.02.28.529746
Peer Review
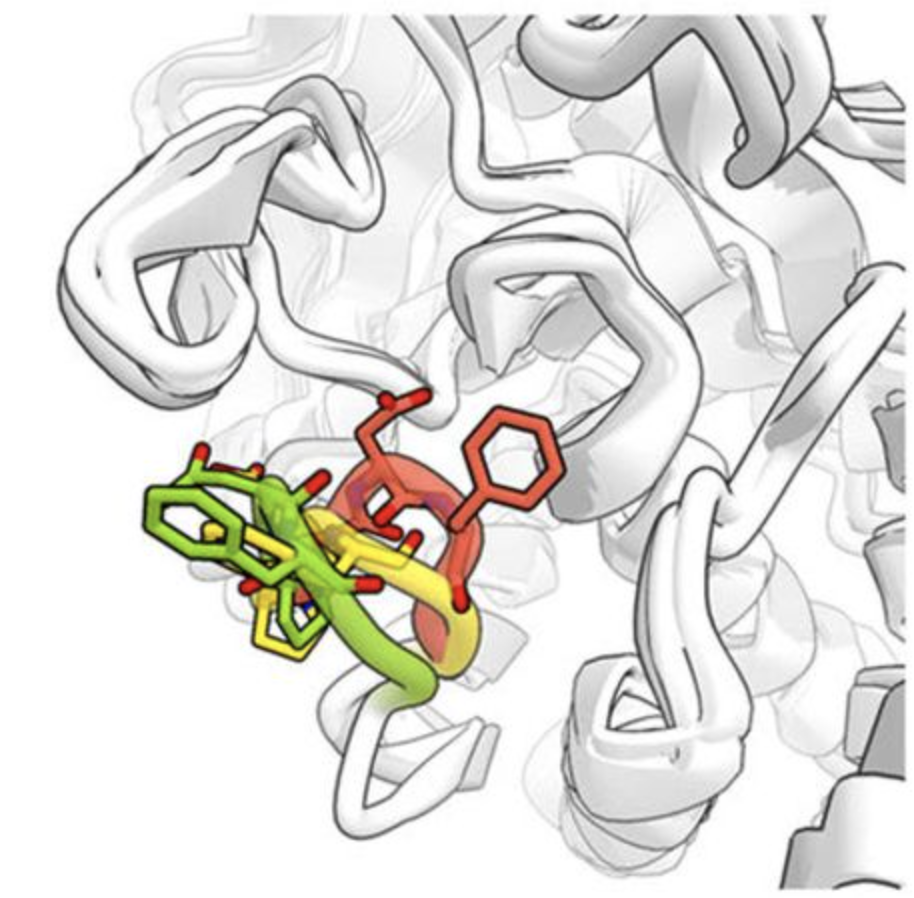
Discovery and validation of the binding poses of allosteric fragment hits to PTP1b: From molecular dynamics simulations to X-ray crystallography.
Greisman JB, Willmore L, Yeh CY, Giordanetto F, Shahamadtar S, Nisonoff H, Maragakis P, Shaw DE.
Reviewed by: San Felipe CJ, Fraser J.
- Biorxiv Preprint v2: 516467
- Review: 2tiiqzd
- PREreview.org: doi-10.1101-2022.11.14.516467
Peer Review
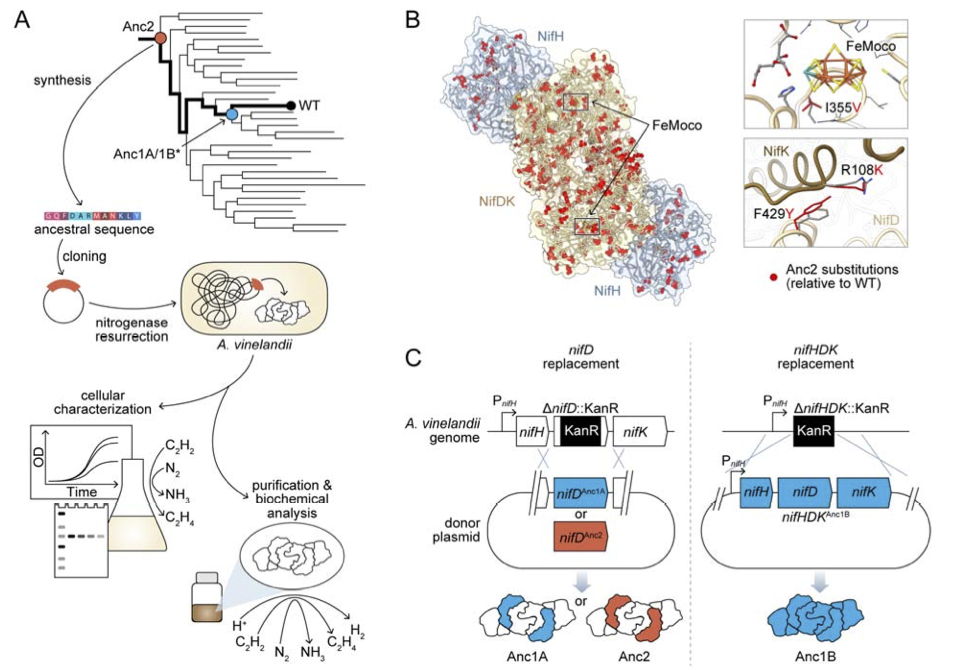
Nitrogenase resurrection and the evolution of a singular enzymatic mechanism.
Garcia AK, Harris DF, Rivier AJ, Carruthers BM, Pinochet-Barros A, Seefeldt L, Kaçar B.
Reviewed by: Macdonald CB, Lee S.
The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan T, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, and Clapham DE.
Reviewed by: Anonymous Reviewer, Fraser JS.
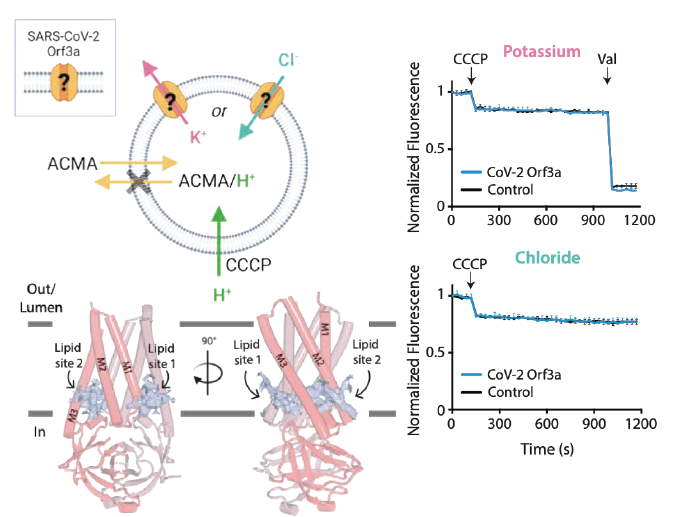
- Biorxiv Preprint v1: 506428
- Review: 2t15ojf
- Explanatory tweetstorm by James Fraser
Peer Review
Additional Link
- PMID: 36695574
Post-Review
Access the paper
Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Mehlman TS, Biel JT, Azeem SM, Nelson ER, Hossain S, Dunnett LE, Paterson NG, Douangamath A, Talon R, Axford D, Orins H, von Delft F, Keedy DA.
Reviewed by: San Felipe CJ, Asthana P, Fraser J.
Activating alternative transport modes in a multidrug resistance efflux pump to confer chemical susceptibility.
Spreacker PJ, Thomas N, Beeninga WF, Brousseau M, Porter CJ, Hibbs KM, Henzler-Wildman KA.
Reviewed by: Macdonald CB.
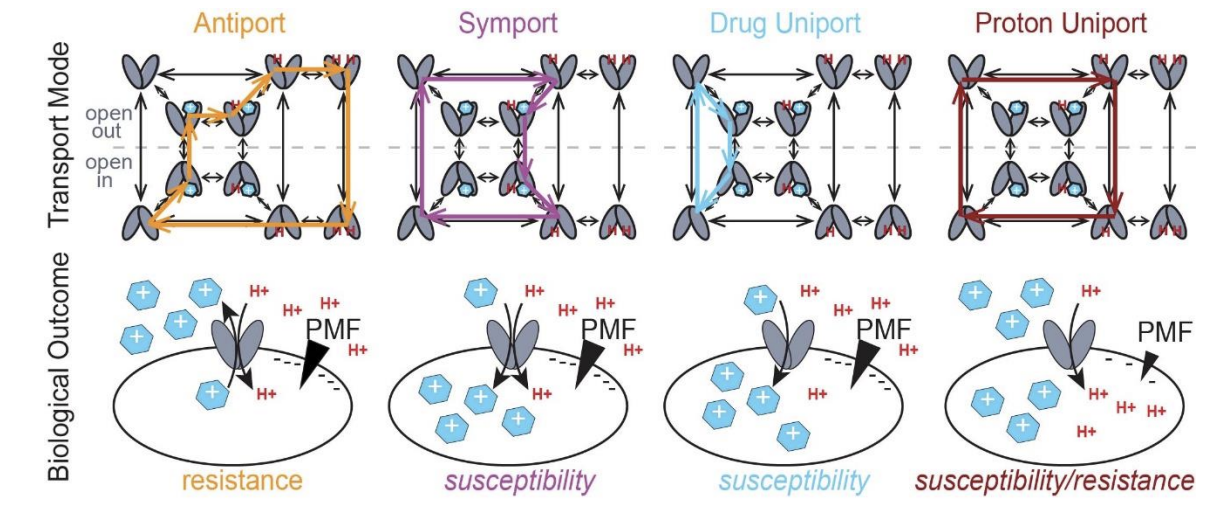
- Biorxiv Preprint v2: 471113
- PREreview.org: doi-10.1101-2021.12.04.471113
Peer Review
Torsion angles to map and visualize the conformational space of a protein.
Ginn HM.
Reviewed by: Ravikumar A, Fraser JS.
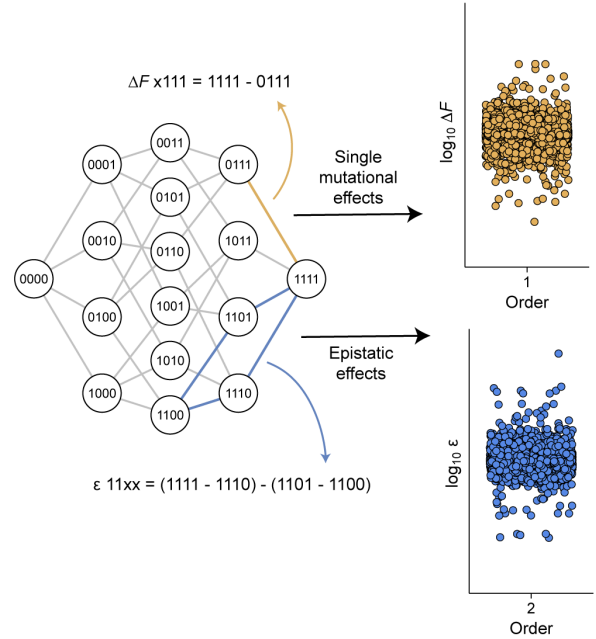
Higher-order epistasis creates idiosyncrasy, confounding predictions in protein evolution.
Buda K, Miton CM, Tokuriki N.
Reviewed by: Macdonald CB, Lee S, Fraser JS.
- Biorxiv Preprint v1: 505194
- PREreview.org: doi-10.1101-2022.09.07.505194
Peer Review
- PMID: 38129396
Post-Review
Access the paper
Cryo-EM structure and B-factor refinement with ensemble representation.
Cragnolini T, Beton J, Topf M.
Reviewed by: Wankowicz S, Fraser JS.
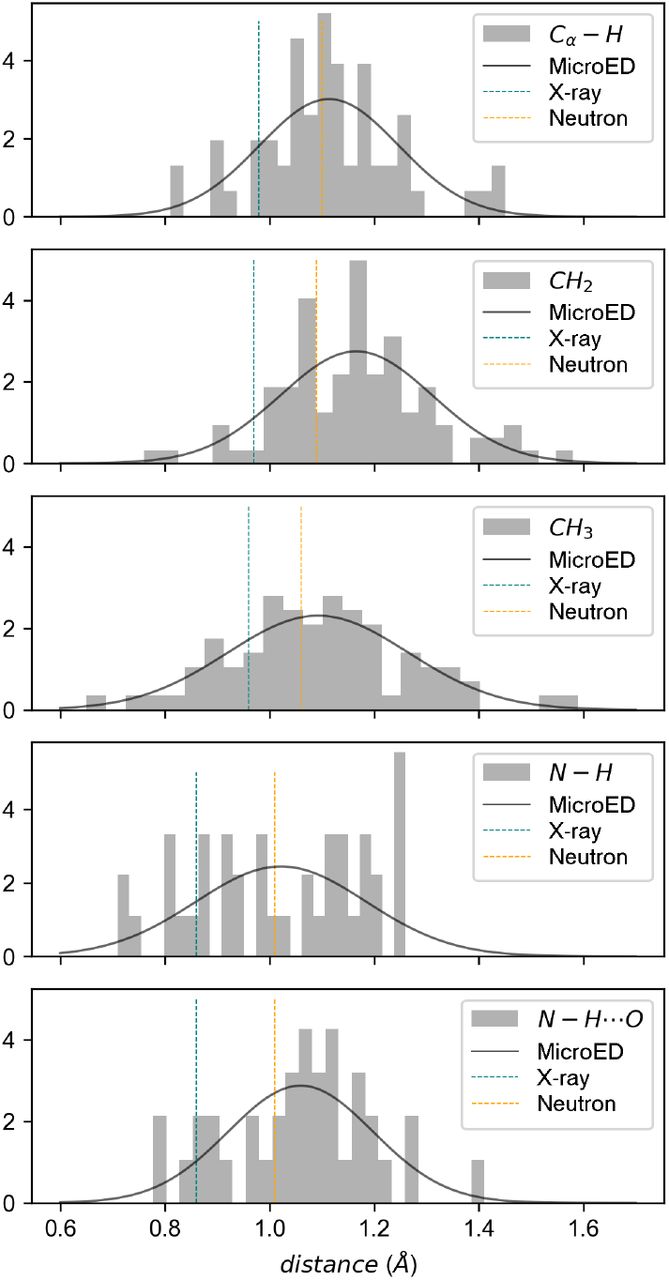
Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
Clabbers MTB, Martynowycz MW, Hattne J, Gonen T.
Reviewed by: Asthana P, Fraser JS.
The evolutionary history of class I aminoacyl-tRNA synthetases indicates early statistical translation.
Jabłońska J, Chun-Chen Y, Longo LM, Tawfik DS, Gruic-Sovulj I.
Reviewed by: Ravikumar A, Fraser JS.
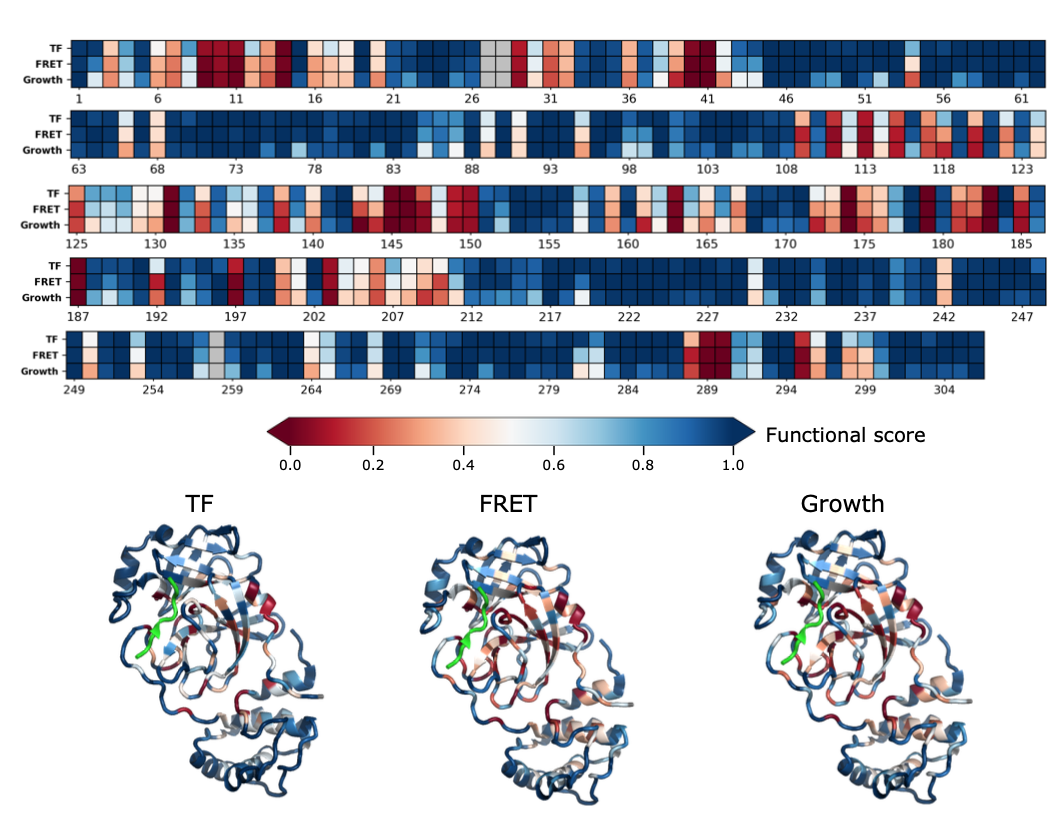
Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms.
Flynn JM, Samant N, Schneider-Nachum G, Barkan DT, Yilmaz NK, Schiffer CA, Moquin SA, Dovala D, Bolon DNA.
Reviewed by: Macdonald CB, Fraser JS.
Ab initio phasing macromolecular structures using electron-counted MicroED data.
Martynowycz MW, Clabbers MTB, Hattne J, and Gonen T.
Reviewed by: Young ID, Fraser JS.
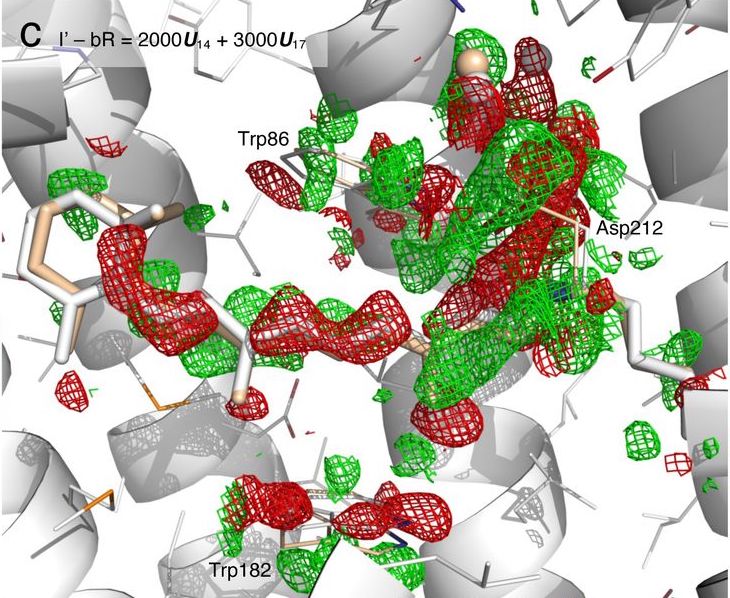
Photoinduced Isomerization Sampling of Retinal in Bacteriorhodopsin.
Ren Z.
Reviewed by: Wolff A, Ravikumar A, Fraser JS.
Sequence- and chemical specificity define the functional landscape of intrinsically disordered regions.
Langstein-Skora I, Schmid A, Emenecker RJ, Richardson MOG, Götz MJ, Payer SK, Korber P, Holehouse AS.
Reviewed by: Chen D, San Felipe CJ, Fraser JS.
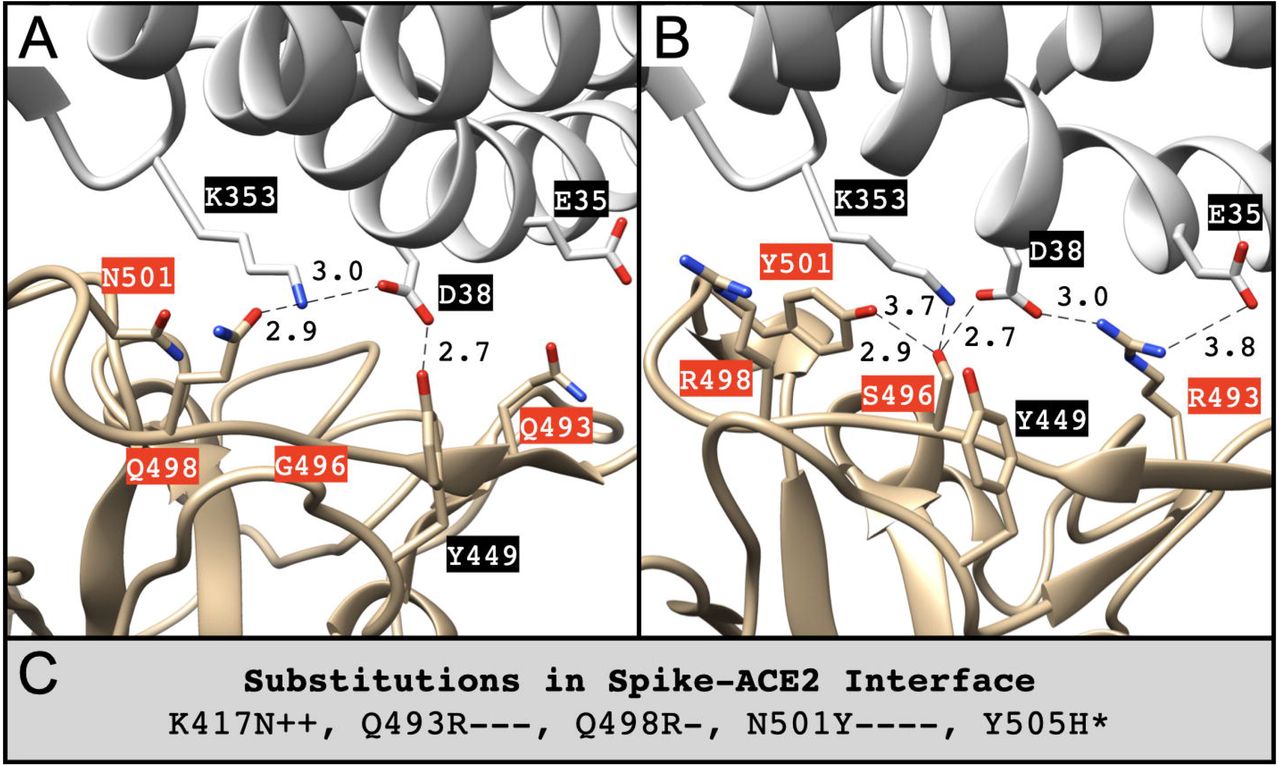
Structural models of SARS-CoV-2 Omicron variant in complex with ACE2 receptor or antibodies suggest altered binding interfaces.
Lubin JH, Markosian C, Balamurugan D, Pasqualini R, Arap W, Burley SK, and Khare SD.
Reviewed by: Díaz RE, Fraser JS.
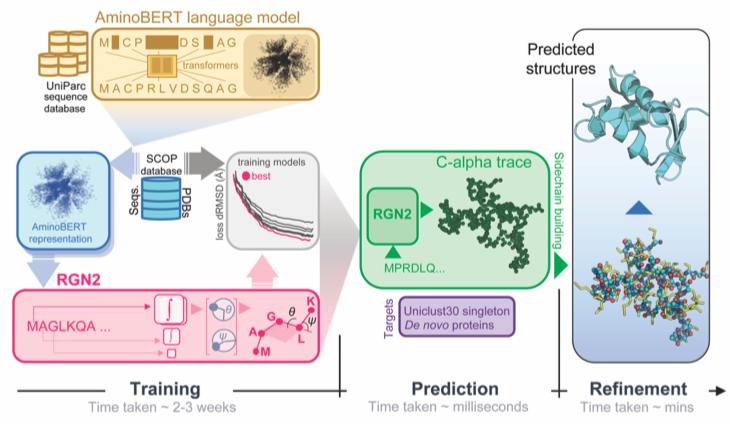
Single-sequence protein structure prediction using language models from deep learning.
Chowdhury R, Bouatta N, Biswas S, Rochereau C, Church GM, Sorger PK, and AlQuraishi M.
Reviewed by: Madani A, Fraser JS.
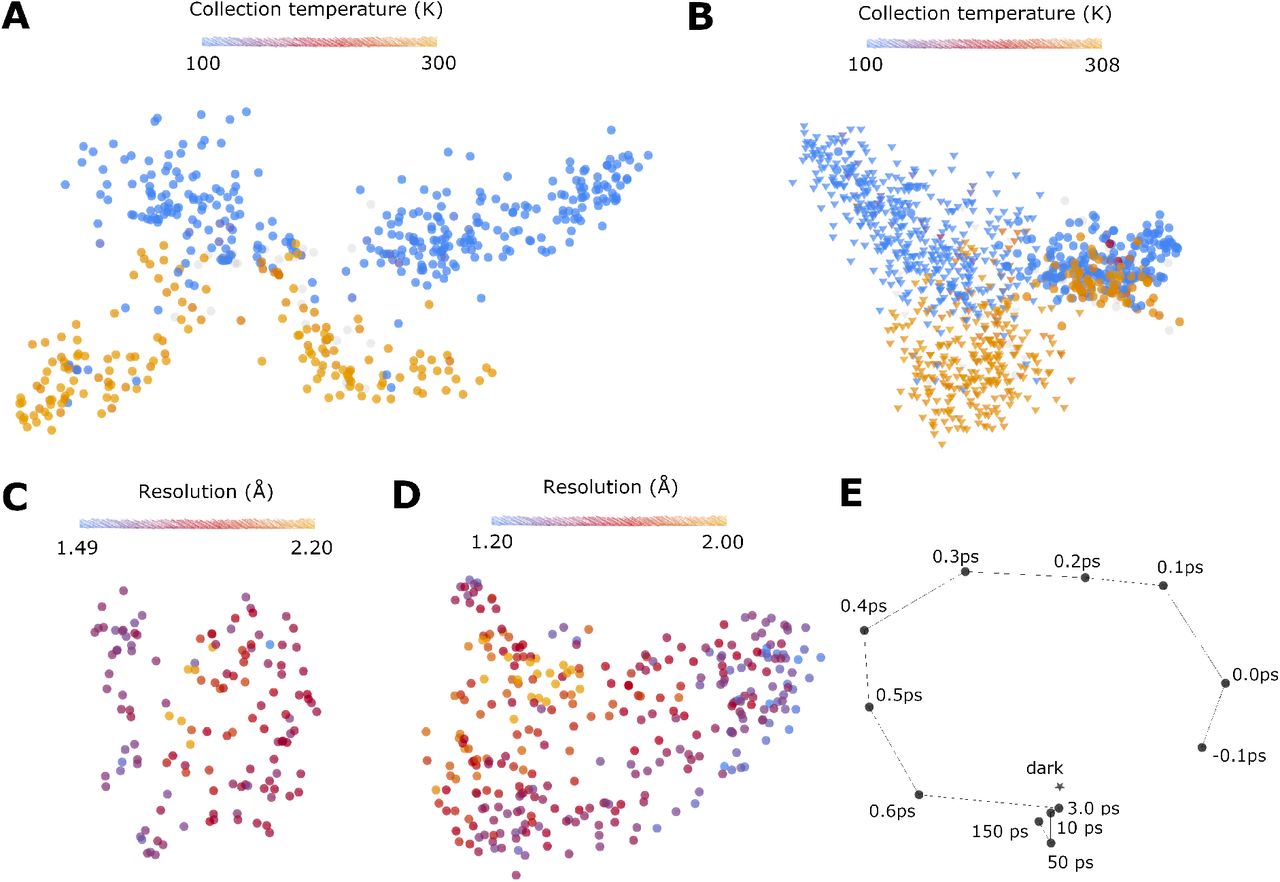
Effects of cryo-EM cooling on structural ensembles.
Bock LV, and Grubmüller H.
Reviewed by: Young ID, Fraser JS.

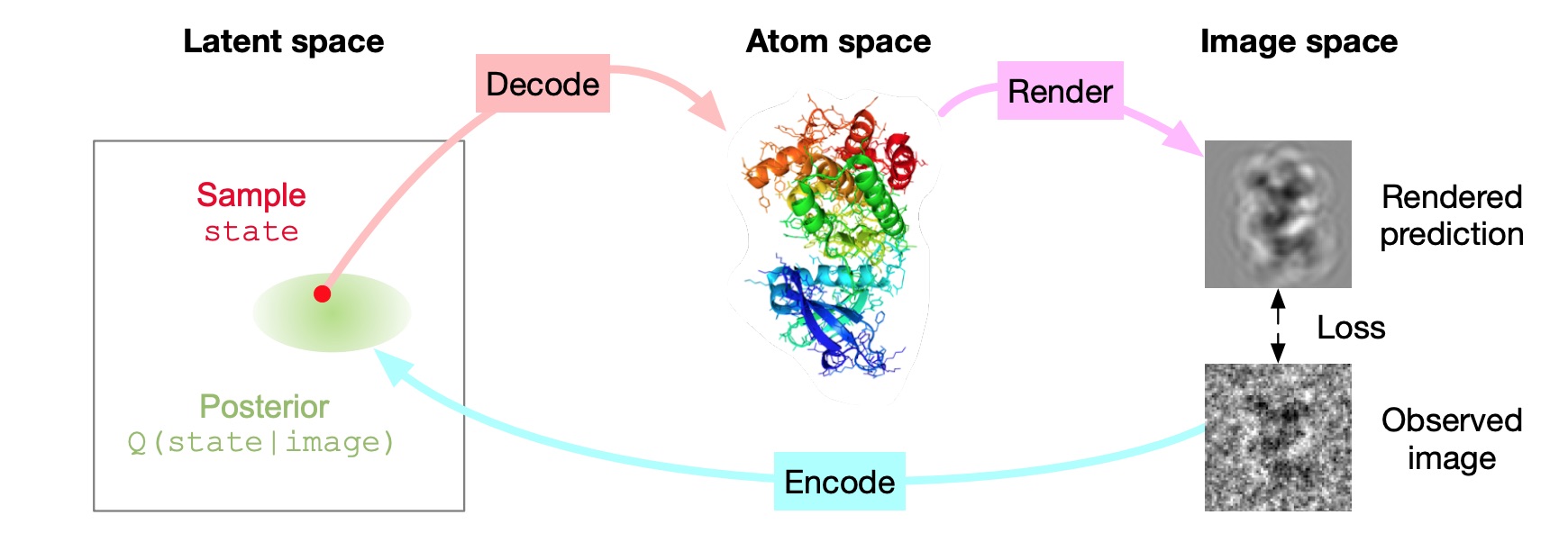
Inferring a Continuous Distribution of Atom Coordinates from Cryo-EM Images using VAEs.
Rosenbaum D, Garnelo M, Zielinski M, Beattie C, Clancy E, Huber A, Kohli P, Senior AW, Jumper J, Doersch C, Eslami SMA, Ronneberger O, and Adler J.
Reviewed by: Wankowicz SA, Young ID, Asarnow D, Fraser JS.
reciprocalspaceship: a Python library for crystallographic data analysis.
Greisman JB, Dalton KM, and Hekstra DR.
Reviewed by: Young ID.
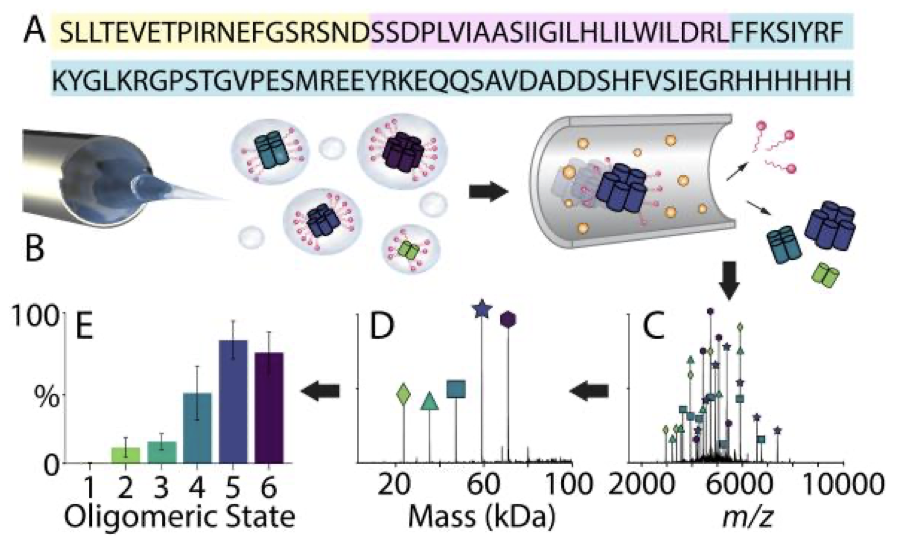
Influenza A M2 Channel Oligomerization is Sensitive to its Chemical Environment.
Townsend JA, Sanders HM, Rolland AD, Prell JS, Wang J, and Marty MT.
Reviewed by: Fraser JS, Anonymous Reviewer #1, Anonymous Reviewer #2, Anonymous Reviewer #3, Anonymous Reviewer #4.
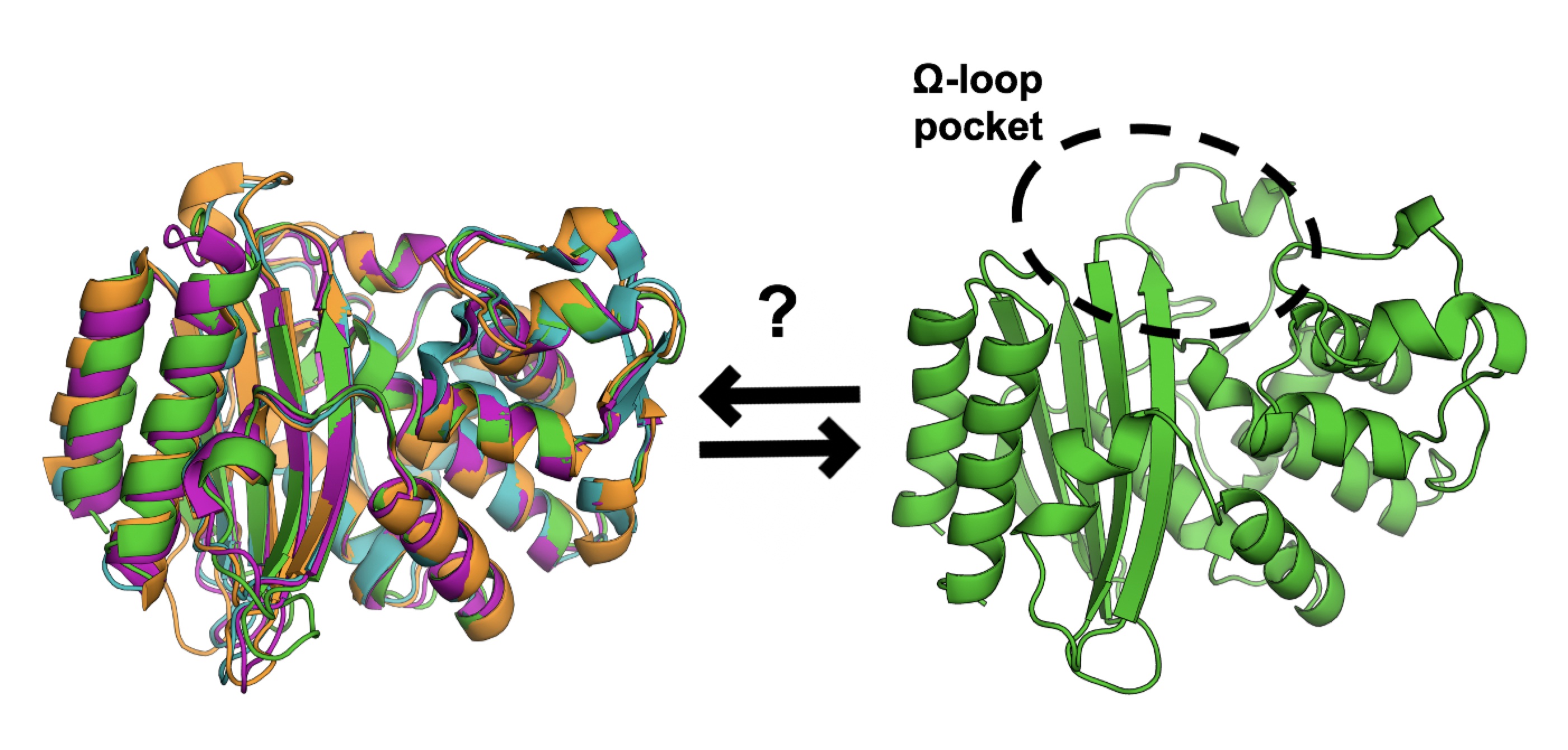
Opening of a Cryptic Pocket in β-lactamase Increases Penicillinase Activity.
Knoverek CR, Mallimadugula UL, Singh S, Rennella E, Frederick TE, Yuwen T, Raavicharla S, Kay LE, and Bowman GR.
Reviewed by: Coyote-Maestas W, Díaz RE, Fraser JS.
Mapping the Functional Landscape of the Receptor Binding Domain of T7 Bacteriophage by Deep Mutational Scanning.
Huss P, Meger A, Leander M, Nishikawa K, and Raman S.
Reviewed by: Coyote-Maestas W, Fraser JS.
Improving SARS-CoV-2 structures: Peer review by early coordinate release.
Croll TI, Williams CJ, Chen VB, Richardson DC, and Richardson JS.
Reviewed by: Young ID, Fraser JS.
Croll and coworkers describe a unique form of peer-review enabled by the early release of biomolecular structure coordinates and density maps and detail its critical role in the search for SARS-CoV-2 vaccines and treatments.

- Zenodo Record: 4697901
Peer Review
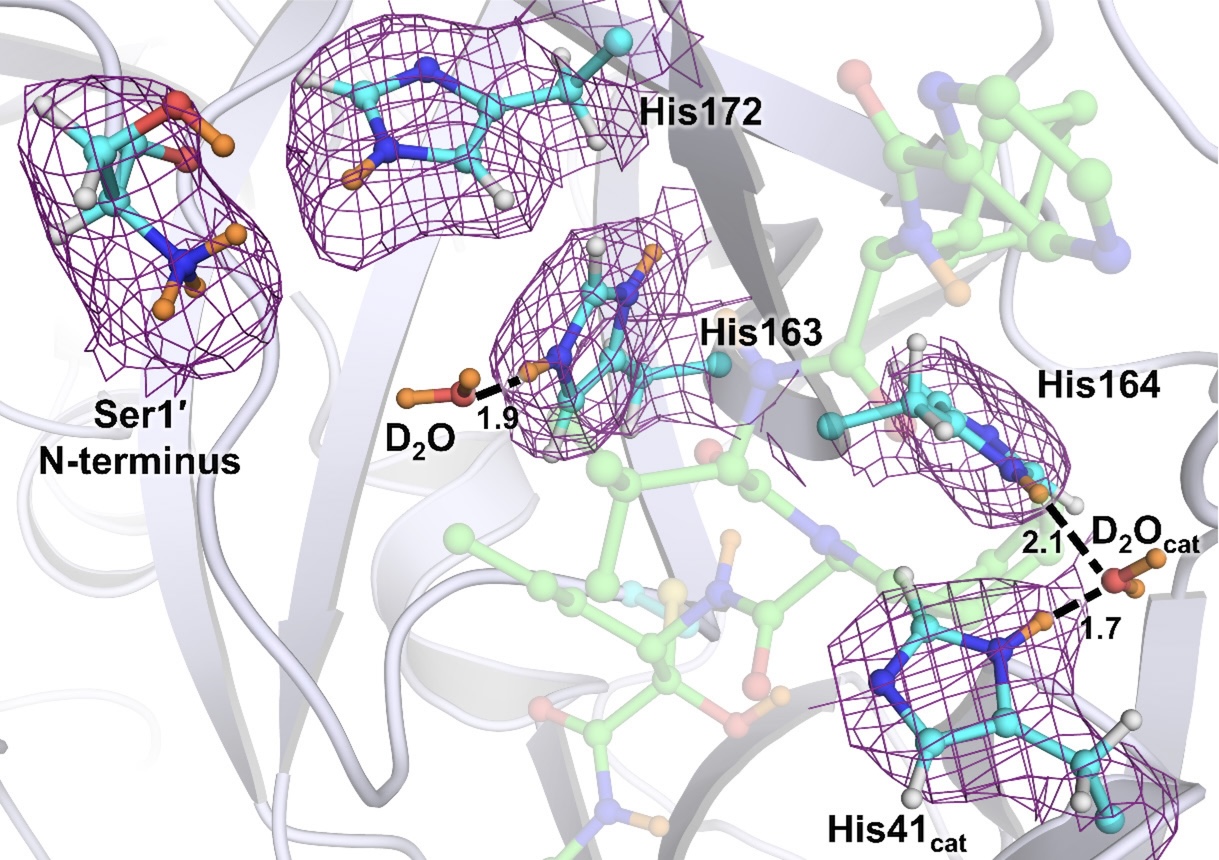
Inhibitor Binding Modulates Protonation States in the Active Site of SARS-CoV-2 Main Protease.
Kneller DW, Phillips G, Weiss KL, Zhang Q, Coates L, and Kovalevsky A.
Reviewed by: Correy G, Fraser JS.
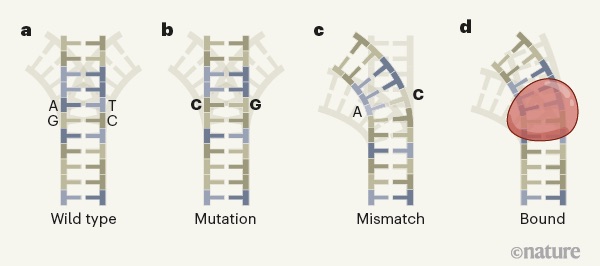
DNA mismatches reveal conformational penalties in protein–DNA recognition.
Afek A, Shi H, Rangadurai A, Sahay H, Senitzki A, Xhani S, Fang M, Salinas R, Mielko Z, Pufall MA, Poon GMK, Haran TE, Schumacher MA, Al-Hashimi HM, and Gordân R.
Reviewed by: Kundert K, Fraser JS.
- Biorxiv Preprint v1: 705558
Peer Review
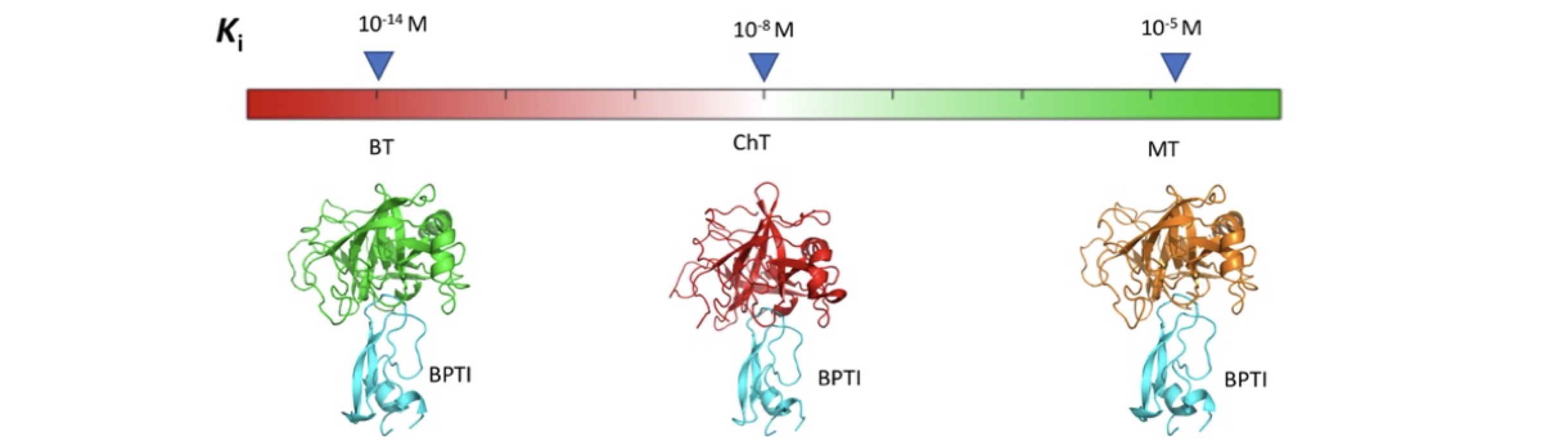
Climbing up and down binding landscapes: a high-throughput study of mutational effects in homologous protein-protein complexes.
Heyne M, Shirian J, Cohen I, Peleg Y, Radisky ES, Papo N, and Shifman JM.
Reviewed by: Estevam G, Fraser JS.
Making the invisible enemy visible.
Croll T, Diederichs K, Fischer F, Fyfe C, Gao Y, Horrell S, Joseph AP, Kandler L, Kippes O, Kirsten F, Müller K, Nolte K, Payne A, Reeves MG, Richardson J, Santoni G, Stäb S, Tronrud D, Williams C, and Thorn A.
Reviewed by: Young ID, Fraser JS.
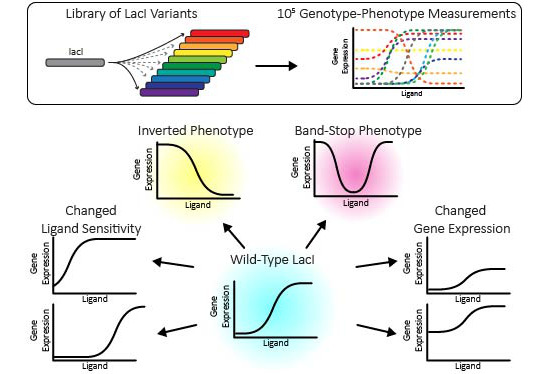
The genotype-phenotype landscape of an allosteric protein.
Tack DS, Tonner PD, Pressman A, Olson ND, Levy SF, Romantseva EF, Alperovich N, Vasilyeva O, and Ross D.
Reviewed by: Coyote-Maestas W, Fraser JS.
MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Martynowycz MW, Shiriaeva A, Ge X, Hattne J, Nannenga BL, Cherezov V, and Gonen T.
Reviewed by: Young ID, Fraser JS.
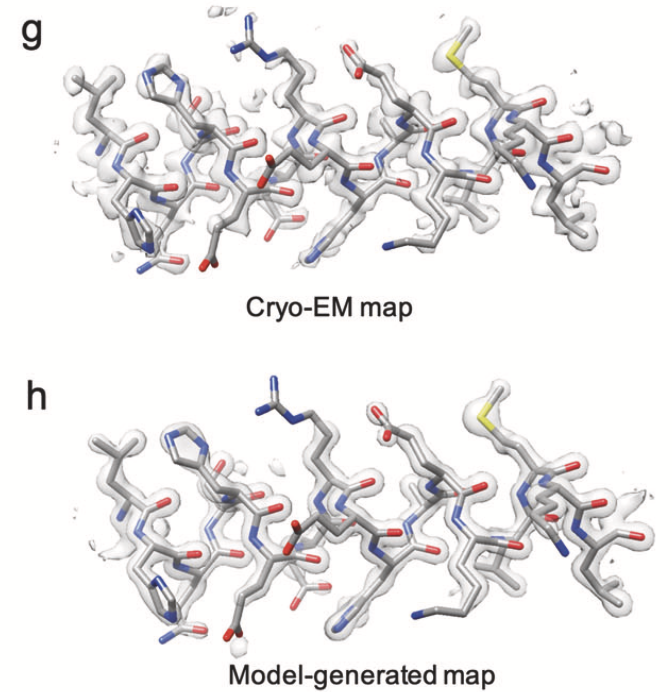
Resolving Individual-Atom of Protein Complex using Commonly Available 300-kV Cryo-electron Microscopes.
Zhang K, Pintilie GD, Li S, Schmid MF, and Chiu W.
Reviewed by: Wankowicz S, Fraser JS.
Near-Physiological-Temperature Serial Femtosecond X-ray Crystallography Reveals Novel Conformations of SARS-CoV-2 Main Protease Active Site for Improved Drug Repurposing.
Durdagi S, Dağ C, Dogan B, Yigin M, Avsar T, Buyukdag C, Erol I, Ertem B, Calis S, Yildirim G, Orhan MD, Guven O, Aksoydan B, Destan E, Sahin K, Besler SO, Oktay L, Shafiei A, Tolu I, Ayan E, Yuksel B, Peksen AB, Gocenler O, Yucel AD, Can O, Ozabrahamyan S, Olkan A, Erdemoglu E, Aksit F, Tanisali G, Yefanov OM, Barty A, Tolstikova A, Ketawala GK, Botha S, Dao EH, Hayes B, Liang M, Seaberg MH, Hunter MS, Batyuk A, Mariani V, Su Z, Poitevin F, Yoon CH, Kupitz C, Sierra RG, Snell E, and DeMirci H.
Reviewed by: Correy G, Fraser JS.
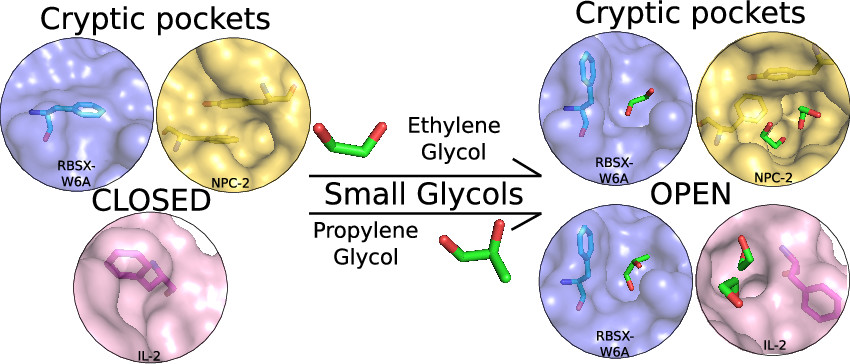
Small Glycols Discover Cryptic Pockets on Proteins for Fragment-based Approaches.
Bansia H, Mahanta P, Yennawar NH, and Ramakumar S.
Reviewed by: Díaz RE, Fraser JS.
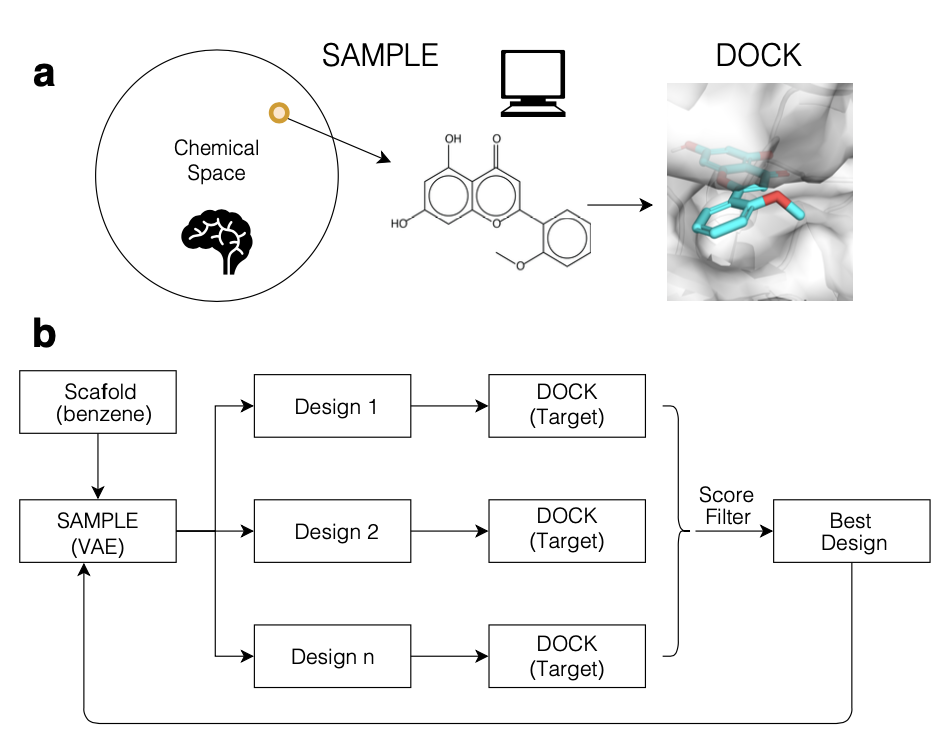
Navigating Chemical Space By Interfacing Generative Artificial Intelligence and Molecular Docking.
Xu Z, Wauchope O, and Frank AT.
Reviewed by: Fraser JS.
An RNA dynamic ensemble at atomic resolution.
Shi H, Rangadurai A, Assi HA, Roy R, Case DA, Herschlag D, Yesselman JD, and Al-Hashimi HM.
Reviewed by: Fraser JS.
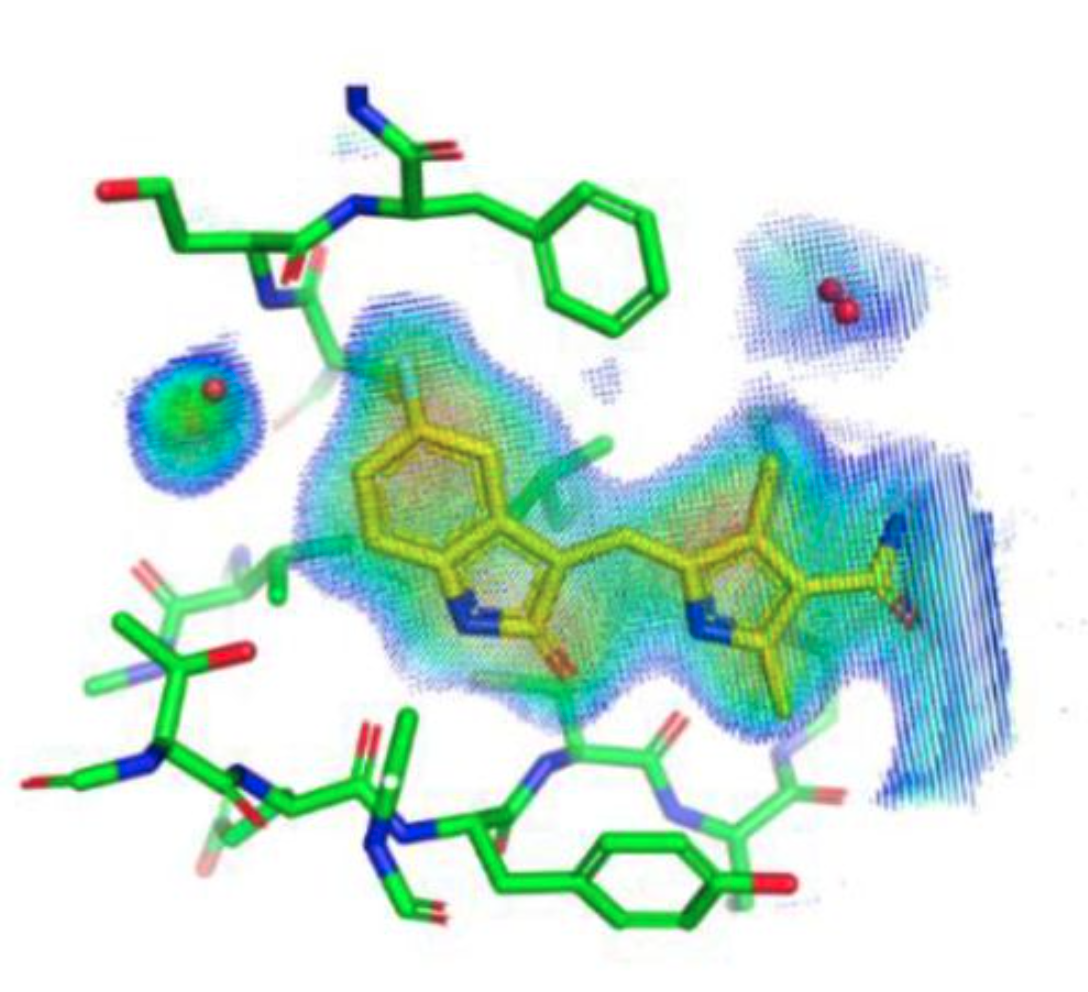
Using a fragment-based approach to identify novel chemical scaffolds targeting the dihydrofolate reductase (DHFR) from Mycobacterium tuberculosis.
Ribeiro JA, Hammer A, Zúñiga GAL, Chavez-Pacheco SM, Tyrakis P, de Oliveira GS, Kirkman T, Bakali JE, Rocco SA, Sforça ML, Parise-Filho R, Coyne AG, Blundell TL, Abell C, and Dias MVB.
Reviewed by: Díaz RE, Chaires HA, Fraser JS.
Improvement of cryo-EM maps by density modification.
Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, and Afonine PV.
Reviewed by: Young ID, Rohou A, Fraser JS.
Functional Plasticity and Evolutionary Adaptation of Allosteric Regulation.
Leander M, Yuan Y, Meger A, Cui Q, and Raman S.
Reviewed by: Coyote-Maestas W, Fraser JS.
Discovery of a cryptic allosteric site in Ebola’s ‘undruggable’ VP35 protein using simulations and experiments.
Cruz MA, Frederick TE, Singh S, Vithani N, Zimmerman MI, Porter JR, Moeder KE, Amarasinghe GK, and Bowman GR.
Reviewed by: Pellegrino J, Fraser JS.
Structural and Functional Characterization of G Protein-Coupled Receptors with Deep Mutational Scanning.
Jones EM, Lubock NB, Venkatakrishnan AJ, Wang J, Tseng AM, Paggi JM, Latorraca NR, Cancilla D, Satyadi M, Davis JE, Babu MM, Dror RO, and Kosuri S.
Reviewed by: Estevam G, Fraser JS.
Automatic building of protein atomic models from cryo-EM density maps using residue co-evolution.
Bouvier G, Bardiaux B, Pellarin R, Rapisarda C, and Nilges M.
Reviewed by: Díaz RE, Young ID, Fraser JS.
Diffuse X-ray Scattering from Correlated Motions in a Protein Crystal.
Meisburger SP, Case DA, and Ando N.
Reviewed by: Wolff A, Fraser JS.
Improved chemistry restraints for crystallographic refinement by integrating the Amber force field into Phenix.
Moriarty NW, Janowski PA, Swails JM, Nguyen H, Richardson JS, Case DA, and Adams PD.
Reviewed by: Wankowicz S, Pierce L, Fraser JS.
Measurement of Atom Resolvability in CryoEM Maps with Q-scores.
Pintilie G, Zhang K, Su Z, Li S, Schmid MF, and Chiu W.
Reviewed by: Barad B, Fraser JS.
Stimulus-responsive self-assembly of protein-based fractals by computational design.
Hernández NE, Hansen WA, Zhu D, Shea ME, Khalid M, Manichev V, Putnins M, Chen M, Dodge AG, Yang L, Marrero-Berríos I, Banal M, Rechani P, Gustafsson T, Feldman LC, Lee SH, Wackett LP, Dai W, and Khare SD.
Reviewed by: Young ID, Fraser JS.
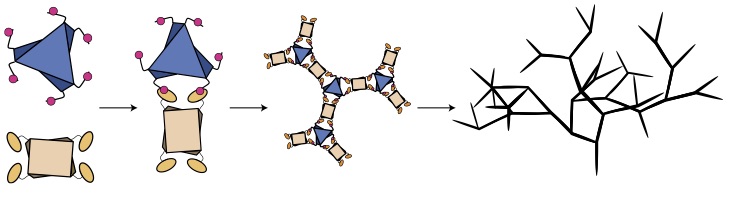
A microtubule RELION-based pipeline for cryo-EM image processing.
Cook AD, Manka SW, Wang S, Moores CA, and Atherton J.
Reviewed by: Young ID, Fraser JS.
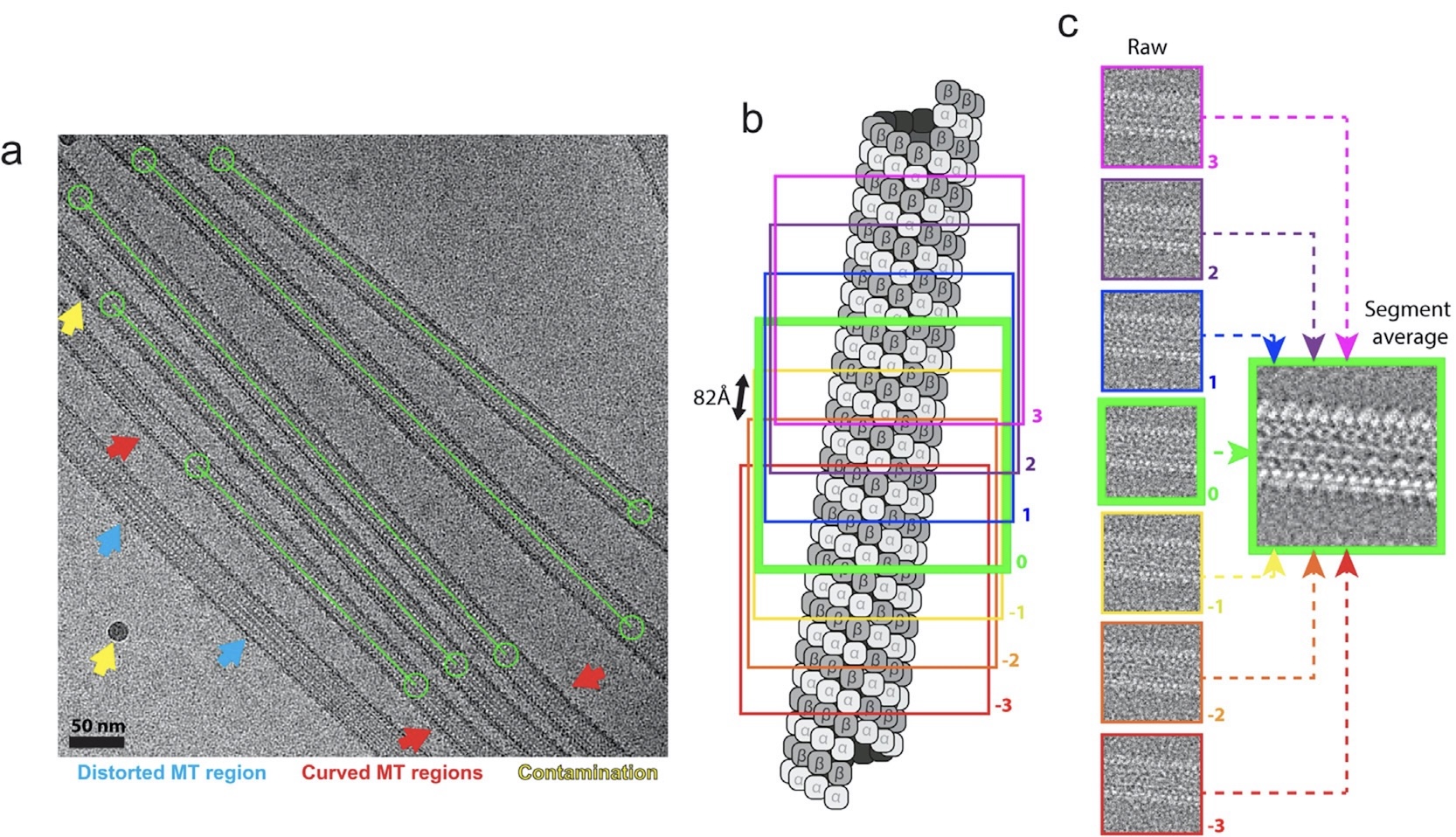
Identifying dynamic, partially occupied residues using anomalous scattering.
Rocchio S, Duman R, Omari KE, Mykhaylyk V, Yan Z, Wagner A, Bardwell JCA, and Horowitz S.
Reviewed by: Correy G, Fraser JS.
A chemical interpretation of protein electron density maps in the worldwide protein data bank.
Yao S, and Moseley HNB.
Reviewed by: Díaz RE, Fraser JS.
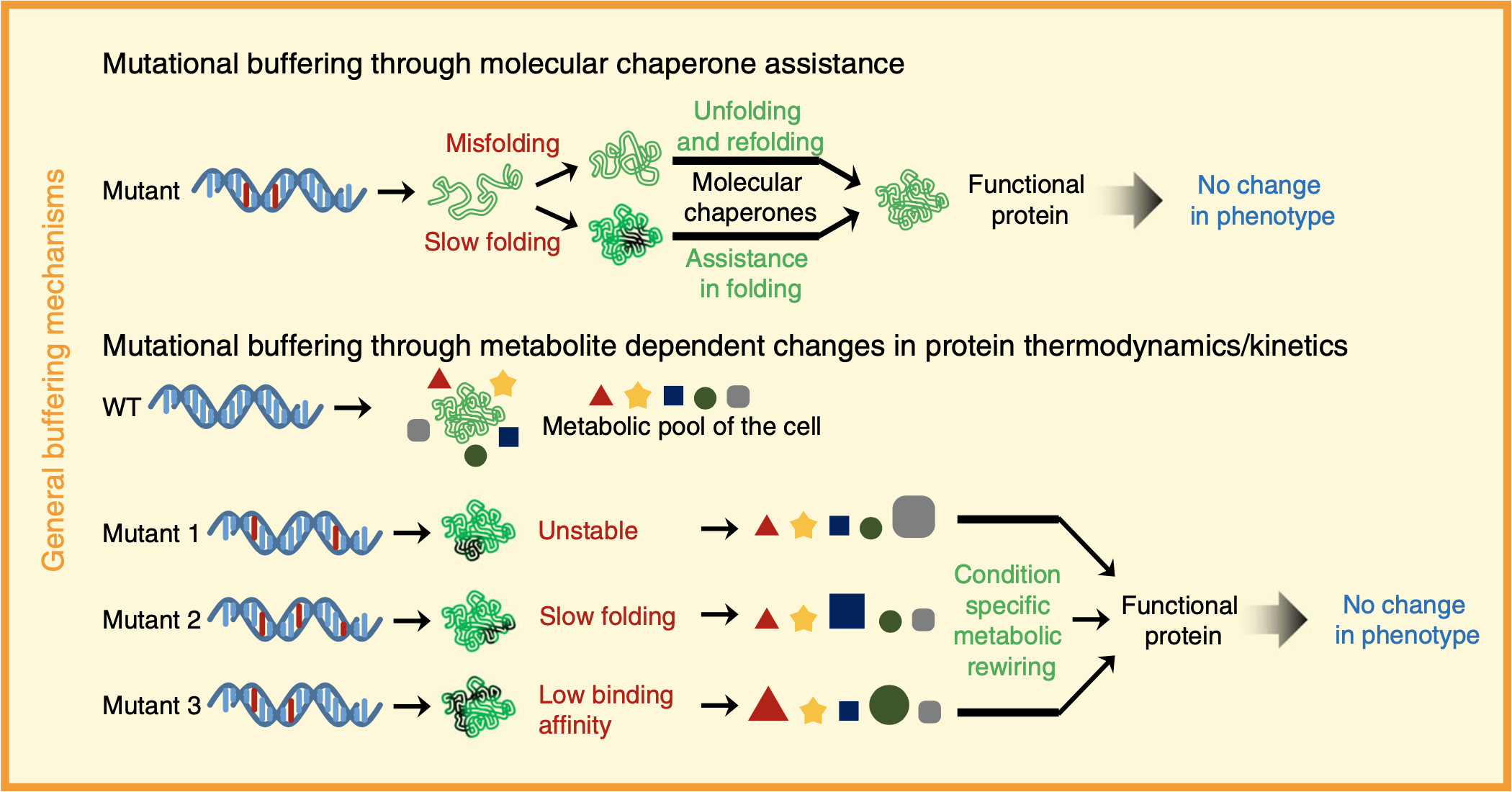
Distinct metabolic states of a cell guide alternate fates of mutational buffering through altered proteostasis.
Verma K, Saxena K, Donaka R, Chaphalkar A, Rai MK, Shukla A, Zaidi Z, Dandage R, Shanmugam D, and Chakraborty K.
Reviewed by: Thompson E, Fraser JS.
Collection of continuous rotation MicroED Data from Ion Beam Milled Crystals of Any Size.
Martynowycz MW, Zhao W, Hattne J, Jensen GJ, and Gonen T.
Reviewed by: Fraser JS.
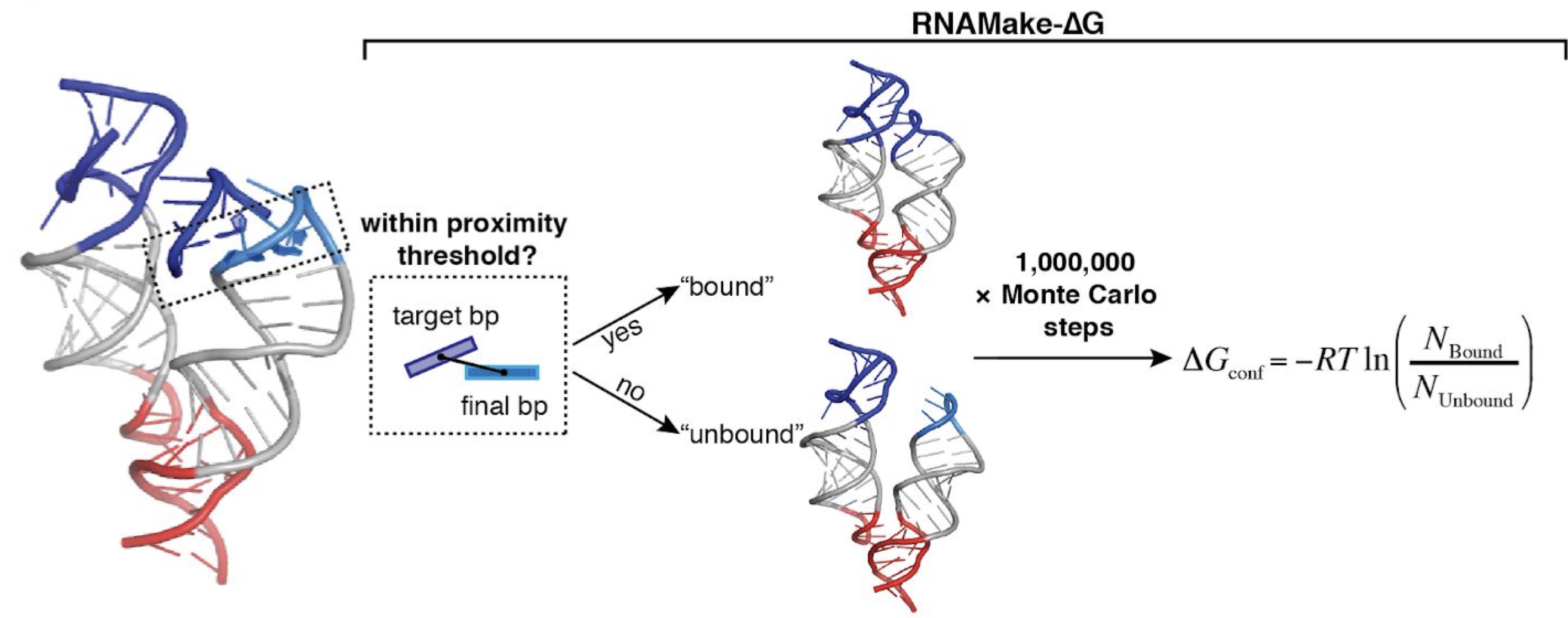
RNA tertiary structure energetics predicted by an ensemble model of the RNA double helix.
Yesselman JD, Denny SK, Bisaria N, Herschlag D, Greenleaf WJ, and Das R.
Reviewed by: Fraser JS.
Accelerating Scientific Publication in Biology.
Vale R.
Reviewed by: Fraser JS.