James Fraser
29 September 2023
tags:
#seminar
Longtime friend of the lab, Michael Wall, will be visiting and deliver a seminar on his pioneering work using diffuse X-ray scattering and molecular dynamics to study proteins.
November 2nd, 2023 4:00pm in GH S201
Diffuse X-Ray Scattering to Shed Light on Protein Dynamics
Michael Wall, Los Alamos National Laboratory
Dynamics in protein crystals gives rise to diffuse X-ray scattering – intensity beneath and between the Bragg peaks in diffraction experiments. Recent improvements in X-ray beamlines and detectors have created new opportunities for using diffuse scattering to understand protein dynamics. In this talk I will introduce some basic concepts about diffuse scattering from protein crystals and the connection to dynamics. I also will review some modern approaches to diffuse data collection, processing, analysis, modeling, and simulation.

Stephanie Wankowicz
09 August 2023
tags:
#deij_jc
Background
Our journal clubs aim to provide an environment for continued learning and critical discussion. Based on the discussion, we also brainstorm action items that individuals and labs can implement. Our discussions and proposed interventions reflect our opinions based on our identities and lived experiences. Consequently, they may differ from the discussions held by those with other identities and/or experiences.
This journal club took place among the entire Fraser lab. Due to the size, we split into three groups. Each group had unique but overlapping conversations. Below are the major points discussed by each group.
Discussion Leader: Stephanie Wankowicz, Daphne Chen, Eric Greene
Articles:
“Differential retention contributes to racial/ethnic disparity in U.S. academia”
Summary and Key Points:
The top ranks of academia, particularly tenured faculty positions, suffer from a glaring lack of racial diversity(1). The cause of this lack of diversity is commonly attributed to challenges in recruitment and retention. Recruitment involves increasing enrollment of students in undergraduate or graduate programs, while retention focuses on keeping people in the ‘academic pipeline’ as they transition from role to role. Insufficient recruitment is widely recognized as a critical contributor to the lack of diversity in STEM fields; however, retention also significantly contributes to this disparity (2-4).
This paper addresses these concerns by focusing on differential retention. They frame retention through a null model, which states that if all else was equal, given the number of academics at stage i in a particular race category, there should be a proportional number of academics in that race category at stage i + 1. They examine each NIH race category’s academic career trajectory trends (5). The authors then compare the distribution predicted by this model to what is observed in NSF survey data. This comparison allows them to ask at which stage each race tends to “fall out” of the academic pipeline. The trends presented in this paper represent a significant dropout of certain races when moving from one stage to another. This was most evident from the grad school to postdoc stage, with a significant dropout of Black and Hispanic academics.
Racial Categories:
This study utilizes the NIH racial categories, which are extremely broad. We discussed how these categories oversimplify racial groups in the United States. The international scholars in the lab also provided perspective on how racial categories are a region- or country-specific issue, with many countries not discussing the issue of race due to a much more homogenous society.
Academic Career Trajectories:
The study broke down the transition from each ‘stage’ of an academic career (graduate, post-doc, pre-tenure faculty, post-tenure faculty). While this analysis removes many confounding factors, the academic career path is not for everyone, and performing this analysis with only those who want to enter the next stage of academia may highlight differences.
However, given this paper’s clear trends and the lack of survey data on career goals, adding this category will likely remain the conclusion. While racism is at the core of these disparities, we discussed specific differences at each career stage and potential solutions.
We also discussed the relative need for more career guidance support for people in the postdoc phase, including financial and mentorship. University postdoc offices often try to support thousands of trainees with only 1-2 full-time employees (6). This lack of general support isolates trainees, especially considering that many inclusive social groups in graduate programs are not found at the postdoc level (7). This can cause a much more isolating experience.
The lack of funding or support for research projects can be more relevant at the postdoc and pre-tenure phases. Research focused on different racial categories, such as health disparities research, is underfunded (8). Further, there is bias in obtaining funding (9).
Internal Lab Support:
While most of these changes need to occur on an institutional scale, we also acknowledged the significance of peer mentoring in fostering retention and support among lab members. Although being social and building relationships with lab mates can mitigate this feeling, it does not eliminate the sense of not belonging. Recognizing the value of non-lab-related peer mentoring networks, such as connections with individuals from other labs or institutions, we discussed how these external support systems can contribute to a cohesive and well-functioning lab environment. Furthermore, creating a supportive environment can help individual members see their next step in academia and expose them to career options they would not have otherwise considered. We acknowledged the need for regular conversations about careers and the next steps, as many trainees (graduate students and postdocs) tend to put off considering their future to focus on their science, and PIs should be, but are not always, proactive in initiating these conversations.
Hypotheses and potential solutions for improving retention:
We discussed the socioeconomic factors that present significant obstacles for individuals pursuing careers in higher education. These factors include the affordability of college education, wage loss during postdoctoral training, reliance on family support, costs associated with grad school interviews and applications, and the increasing financial burden of each education stage (on top of the cost of moving between educational stages). We also noted that gender plays a prominent role in many of these transitions, especially grad school to postdoc, postdoc to pre-tenure, as this is when many people in the canonical ‘academic pipeline’ have children (with fewer parents pursuing a postdoc, especially with women, as childbearing work falls disproportionately on women) (7). Moreover, the hard work of childbearing falls disproportionately on women, which presents a broad gender-specific barrier to advancement. More detailed data would provide valuable insights into the career trajectories of those opting out of academia, furthering our understanding of the challenges and reasons behind their decisions.
Open Questions:
- How does the dropout rate from one step to another look among only those who desire to continue in academia?
- How different does this trajectory look for different genders?
- Why do people not move on to a postdoc or pre-tenure position?
- There are also significant dropouts observed from pre-tenure to tenured positions. Why are universities not supporting their pre-tenure faculty through the tenure process?
- What impact does observing a historically underrepresented professor not getting tenure have on an institution’s student population?
- We’d love to see follow-up analyses of this data set, particularly how these trends hold up/change for different disciplines and/or institutions. Can we identify and learn from those demonstrating positive progress toward inclusive academic retention?
Proposed Action Items:
- Advocate for increased funding and support of the postdoctoral affairs office.
- Waiving application fees for graduate school and/or University providing travel funding up front (instead of through reimbursement)
- Provide resources or opportunities for trainees to form peer mentoring networks, such as socials/mixers, funding for groups based on specific career/research goals, etc.
- Bringing awareness of these racial disparities to admission committees, hiring committees, and hiring managers.
Citations:
1) Research: Decoupling of the minority PhD talent pool and assistant professor hiring in medical school basic science departments in the US
2) How Gender and Race Stereotypes Impact the Advancement of Scholars in STEM: Professors’ Biased Evaluations of Physics and Biology Post-Doctoral Candidates
3) Academia’s postdoc system is teetering, imperiling efforts to diversify life sciences
4) Tenure Decisions at Southern Cal Strongly Favor White Men, Data in a Rejected Candidate’s Complaint Suggest
5) Racial and Ethnic Categories and Definitions for NIH Diversity Programs and for Other Reporting Purposes
6) Growing Progress in Supporting Postdocs
7) Academia’s postdoc system is teetering, imperiling efforts to diversify life sciences
8) Role of funders in addressing the continued lack of diversity in science and medicine
9) Fraser Lab DEIJ Journal Club - Blinding Grant Peer Review
James Fraser
19 July 2023
As a faculty member at the University of California, San Francisco (UCSF), I am often asked about my approach to evaluating faculty applications. In writing it out, I not only clarify my thinking, but also provide transparency about how one faculty member evaluates applications. Additionally, by sharing this, I hope to get feedback to help improve my own process for evaluating applications in the future.
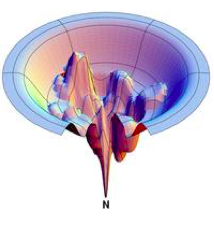
Evaluating faculty applications, in my view, is akin to the process of protein folding, as described by Levinthal’s paradox. Levinthal’s paradox suggests that it would be virtually impossible for a protein to achieve its functional structure by exhaustively exploring every possible conformation due to the sheer number of potential configurations. Instead, proteins navigate through a funnel-like process, where a sequence of favorable local interactions steers the protein toward its final, folded ensemble. When I evaluate faculty applications, I adopt a similar approach. I don’t undertake an exhaustive examination of every single detail of all applications. Instead, I employ a funnel-like process, starting with broader criteria, then progressively narrowing down to more specific aspects of the proposed research program. I strive to do this without resorting to traditional markers of prestige such as the reputation of the journals where they’ve published or their academic pedigree. This process guides me toward the most promising applications that resonate with me both scientifically and in terms of shared scientific values.
The first step in my evaluation process is to review the Diversity, Equity, and Inclusion (DEI) statement. Based on other published rubrics , I assess the applicant’s awareness and involvement in DEI initiatives. I’ll also look over any teaching or mentoring track record as part of this, recognizing that not everyone has had the chance or environment to fully engage in these activities. This is a critical step for me. If an applicant does not demonstrate a strong commitment to DEI, I do not proceed further with their application. This initial screening takes less than five minutes per candidate and typically eliminates about half of the applicants.
Next, I turn my attention to the research statement. The opening page (and especially the opening paragraph!) is crucial here. I look for a clearly articulated problem or a set of problems that the applicant intends to address. If the scientific problem statement, its significance, or the applicant’s approach to solving it are unclear to me, I do not proceed with considering the candidate. This step takes less than two minutes per candidate and usually eliminates another half of the remaining applicants.
For the remaining candidates, I undertake a thorough review of the entire research statement and cover letter. I study the applicant’s key preprints and papers to familiarize myself with their specific scientific questions and approaches. Interestingly, many of the faculty members I’ve been involved in hiring at UCSF had not yet published their major work in a peer-reviewed journal at the time of their application. This is not a deterrent for me; in fact, I embrace preprints wholeheartedly. Preprints provide an open and immediate insight into a researcher’s latest work, and I am fully capable of evaluating them on their own merits. However, what I find less favorable are “confidential manuscripts in review”. Because these do not offer me the same level of transparency as preprints, I won’t review them as part of the application. Including such “confidential manuscripts” demonstrates a disconnect with the open science principles that I value in future colleagues.
During this stage, I also try to evaluate how successful they have been in making progress on key problems in prior career stages by scanning letters of reference and scanning additional papers by the applicant (and often in the field of the applicant).
I also want to clarify what I look for in reference letters, even though they are a minor factor relative to the research proposal and papers of the applicant. It’s common for every applicant to be described as “the best person who has passed through the lab in years,” so overall praise isn’t the differentiator for me. Instead, I focus on three key things:
1 - Context for the scientific barrier the candidate overcame in their prior work.
2 - Discussion of how the candidate’s FUTURE work will differentiate from the thrust of their current lab.
3 - Corroborating data on teaching, mentorship, and outreach.
Letters can add depth to these three dimensions, but rarely detract from them. While it’s not a strict requirement, a well-crafted letter that resonates on these three issues can be immensely helpful in painting a comprehensive picture of the candidate.
This overall step of evaluating the research statement and papers (with a scan of letters of references and other papers) is time-intensive, taking approximately 20 to 40 minutes per candidate. However, this is the point where I decide if a candidate should be evaluated by the entire committee, generally nominating about 10-15 candidates.
At this point, I also get the short list of other members of the committee. Some of my colleagues may weigh other factors such as the prestige of journals where the applicant has published, their academic pedigree, or the likelihood of securing funding. This diversity in evaluation criteria is a strength of a committee approach, provided we are all aware of and acknowledge our biases. We typically get about 100-300 applicants in a cycle, but there is usually a significant overlap in shortlists. Generally, the committee process leads to a shortlist of ~25 candidates.
The next step involves a deeper reflection on each shortlisted application. I spend an additional 30 minutes per application, contemplating the fit of the research statement with our institution and gauging my excitement level about the proposed research. I again consider the DEI and teaching/mentoring efforts. My aim is to identify 5 to 7 applicants that I am extremely enthusiastic about, 10 applicants that I am open to learning more about if other committee members are sufficiently enthusiastic, and 5 to 10 applicants that I am skeptical about but am willing to be convinced by other committee members.
Finally, we (the hiring committee) engage in a comprehensive discussion and ranking process. Each committee member presents their shortlisted candidates, and we collectively rank them for zoom and/or on-site interviews. This process tries to offer a balanced assessment of each candidate, helping us identify the most promising faculty members for UCSF.
In conclusion, my approach to faculty application evaluation is designed to be rigorous and thorough, while being efficient and minimizing proxies of prestige like journal name or institution. I’m cognizant that I have my own implicit and explicit biases, but what is outlined here is a reflection of how I try to identify candidates who not only excel in their research but also share our values. I believe it’s important to share my process, not as a standard, but as an example of one possible approach. I encourage anyone serving on a hiring committee to outline their own unique criteria and detail the process they use to arrive at a shortlist.
Thank you to Prachee Avasthi, Zara Weinberg, Willow Coyote-Maestas, Stephanie Wankowicz, Chuck Sanders, Brian Kelch, and Jeanne Hardy for feedback and discussions about this topic.
Eric Greene
04 November 2022
tags:
#deij_jc
Background
A group of scientists within the Fraser, Coyote-Maestas, and Pinney labs have begun a journal club centered around issues of diversity, equity, inclusion, and justice within academia, specifically in the biological sciences.
Our goal is to provide an environment for continued learning, critical discussion, and brainstorming action items that individuals and labs can implement. Our discussions and proposed interventions reflect our own opinions based on our personal identities and lived experiences, and may differ from the identities and experiences of others. We will recap our discussions and proposed action items through a series of blog posts, and encourage readers to directly engage with DEIJ practitioners and their scholarship to improve your environment.
November 4th, 2022 – Blinding peer review
Discussion Leader: Eric Greene
Articles:
Summary Article: “Funding: Blinding peer review”
Primary Article: “An experimental test of the effects of redacting grant applicant identifiers on peer review outcomes”
Bonus Article: “Strategies for inclusive grantmaking”
Summary and Key Points:
STEM research funding is a highly competitive space that has a persistent lack of diversity and representation, especially at the faculty level. I chose this case study as it discusses one of the largest current racial disparities in STEM, highlights a source of white privilege that directly impacts lab funding, and provides experimental evidence towards one mitigation strategy.
The NIH is a substantial funding source for biomedical research in the US and NIH funding is foundational to the existence of many laboratories that are driving biomedical scientific discovery. However, there is a large and persistent funding gap between White and Black investigators, where Black PIs are funded at 55-60% White PIs rate.
In response to this disparity, the NIH conducted a study on the effects of blinding applicants’ identity and institution on the review of R01 proposals. The goal of this large experiment was to gain an understanding about the role of peer review in facilitating racial bias in grant awards and to understand the extent to which blinding applicant identity could blunt racial bias. The experiment uncovered the following:
-
Scores for applications from Black PIs were unaffected by blinding, but scores for applications from White PIs were significantly lower when the White PIs identity was blinded such that the racial gap was cut in half. This finding could be due to the “Halo effect” where personal/institutional prestige dramatically upweights advantaged/privileged individuals and can be seen as another mechanism fueling a ‘winners keep winning’ phenomena. Indeed the “Halo effect” has been indicated to be a potent factor in manuscript peer-review.
-
The principle critique of invoking the “Halo effect” to rationalize the findings of this study is that proposal writers did not write their proposal with identifying information redacted, it was done administratively with previously reviewed R01 applications, leaving uncertainty regarding the impact of administrative redaction on ‘grantsmanship’. However, we discussed the likelihood that applicants who benefit from individual/institutional prestige would likely write favorably toward this status in their applications thus in effect working to entrench any positive “Halo effect” benefit.
Blinding applicant identification on grant proposals is not a silver bullet that solves racial disparity in NIH funding. Including being imperfect itself, with ~22% of reviewers able to positively identify blinded applicant identity. However, this is one tool that has a demonstrated effect here to blunt reviewer bias. While blinding was somewhat effective here, there are means of double blinding and/or tiered blinding of application materials that can be used instead that may hold greater potential.
A key part of our discussion was about the review criteria for NIH funding that explicitly required a numerical evaluation of the individual and institution. Evaluation of a person contributes to an obligate entanglement of one’s past scientific accomplishments with their future potential during the grant review process. Not only can this equivalence be false (people often can succeed past initial setbacks), but it also can be harmful by promoting an applicant’s self-worth to be tied to their productivity. Funding requires accounting for equipment available to carry out the research, which is important for accountability on the part of the investigator, but does not necessarily require a numerical number. This detailed level of evaluation would prompt reviewers to score prestigious/well-resourced institutions higher even if the same research could be carried out elsewhere. We discussed as an alternative whether equipment/facilities categories could be scored as ‘sufficient’ or ‘insufficient’ and not influence the overall impact score of the application.
Open Questions:
- How does one justly judge an application as fundable?
- The ‘Halo effect’ in consequential academic evaluation processes has amassed supportive evidence beyond grant funding. How do we best de-leverage this effect towards a level playing field?
- Blinding applicant identity can help, even if not perfect, how do we improve blinding processes through an equity lens?
- Another explanation for lower Black PI funding rates stems from the subject matter of study, such as studying health care topics of interest for communities of color, which though important may not necessarily be of high funding value to reviewer or reviewing institute. How can these health care topics be adequately elevated and funded?
- To what extent do non-NIH funding mechanisms also incur racial disparity? What have other organizations tried to mitigate? Have these strategies worked?
Proposed Action Items:
While trainees may have limited influence to change the course of NIH peer review, there are nonetheless actions that one can take:
- Call your Representative/Senators to implore them to raise the NIH budget. The value of NIH sponsored research is high to the general public and with more funds, the 10-30% fund rate will increase and be less demoralizing to independent investigators and trainees.
- Should you find yourself in the position of power as a peer reviewer, practice empathy during the review process and familiarize yourself with bias’ that can crop up in the process
- Vote. The NIH is a government entity and is not immune to political authority figures.
- Encourage unsuccessful applicants to pursue resubmission. Rejection is hard but community can help.
- Encourage other non-federal funding mechanisms to blind reviewers or if they have the budget, to do a study where each application is evaluated blinded and open. Compare the scores and who gets funded.
Galen Correy
08 August 2022
tags:
#how_to
Background
The pan-dataset density analysis (PanDDA) tool developed by Nick Pearce and colleagues at the XChem facility of the Diamond Light Source is a super powerful method for identifying low occupancy states in X-ray crystallography data [1,2]. Why do we care about low occupancy states? For one thing, the field of fragment-based drug discovery relies on tools to identify weakly bound ligands [3,4]. When fragments are soaked into protein crystals, the occupancy of the fragment (i.e. the proportion of protein molecules with a fragment bound) can often be relatively low (e.g. 10-20%). PanDDA helps to identify low occupancy fragments by subtracting the ground-state electron density (i.e. the electron density when no ligand is present) from the changed-state electron density (i.e. the electron density when the ligand is present) [1]. In addition to transforming crystallographic fragment screening, PanDDA can also help to identify and model larger ligands that may bind with relatively high affinity compared to fragments, but still have relatively low occupancy. This discrepancy can arise because ligand occupancy in soaking experiments does not necessarily correlate with binding affinity as measured by solution-based methods. One reason for this is low ligand solubility; it may be difficult to reach 1:1 stoichiometry in a soaking experiment. Another reason is that a binding site may be partially obstructed, or otherwise stabilized in a conformation that decreases the ligand occupancy. The presence of low occupancy states is a fundamental challenge of using crystallographic soaking experiments for determining ligand structures: identifying and resolving these states is the reason that PanDDA is such a powerful method.
PanDDA is a powerful tool for identifying low occupancy states, but it presents crystallographers with a new challenge: actually modeling the states it identifies! The best option is to model both states using alternative occupancy (altloc) identifiers in the coordinate file to distinguish ligand-bound and ligand-free states [1,5] (this results in what we call a multi-state model). However, these multi-state models can be difficult to interpret/visualize, especially for the vast majority of users that are only interested in the ligand-bound state. A related issue is that we want to ensure that users can easily examine the PanDDA event maps that were used to model a ligand. For our recent preprint describing the design and structure-based optimization of ligands targeting the Nsp3 macrodomain, we modeled all the structures using a multi-state approach [6]. We’ve taken the following steps to disseminate the structures and maps as rapidly and helpfully as possible.
-
Multi-state coordinate files and structure factor intensities have been deposited in the PDB (with RELEASE NOW selected)
-
Structure factor intensities in MTZ format, Dimple output, PanDDA event/Z-maps, refined structures and ligand-bound states are available to download from Zenodo
-
Diffraction images are available to download from https://proteindiffraction.org (search by PDB code)
How to extract the ligand-bound state in our multi-state models
Option 1
-
Download coordinates from PDB (e.g. fetch 5SQP in PyMOL)
-
Remove the altloc A coordinates - these correspond to the ligand-free state (remove alt A in PyMOL)
-
The coordinates can then be visualized or saved as a coordinate file (pdb 5SQP_ligand-bound.pdb in PyMOL)
Option 2
-
Use this PyMOL script to fetch the coordinates using the PDB code and extract the ligand-bound state
-
This script removes the altloc records for residues that only have a single conformation modeled in the ligand-bound state and renames the altloc records for residues with multiple conformations (Alternatively: the ligand-bound states can be downloaded directly from Zenodo)
How to inspect PanDDA event maps
Option 1
- Use this script to extract the PanDDA event map from the deposited structure factor CIFs (discussed here)
- The resulting map coefficients in MTZ format can be converted to CCP4 format using phenix.mtz2map.
Option 2
- Download the PanDDA event map in .ccp4 format from Zenodo. (Note: use COOT version 0.8.9.2 to visualize maps.)
Where to next?
Our goal is to use macromolecular structural information to make ligand discovery more efficient. We think that identifying and modeling low occupancy states is critical to this endeavor. Developing automated ways to model the low occupancy states identified by PanDDA is a long-term goal. This will speed up ligand modeling and reduce the error/bias that is often associated with manual approaches.
References
[1] Pearce, N. M., Krojer, T., Bradley, A. R., Collins, P., Nowak, R. P., Talon, R., Marsden, B. D., Kelm, S., Shi, J., Deane, C. M. & von Delft, F. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 8, 15123 (2017).
[2] Schuller, M., Correy, G. J., Gahbauer, S., Fearon, D., Wu, T., Díaz, R. E., Young, I. D., Carvalho Martins, L., Smith, D. H., Schulze-Gahmen, U., Owens, T. W., Deshpande, I., Merz, G. E., Thwin, A. C., Biel, J. T., Peters, J. K., Moritz, M., Herrera, N., Kratochvil, H. T., QCRG Structural Biology Consortium, Aimon, A., Bennett, J. M., Brandao Neto, J., Cohen, A. E., Dias, A., Douangamath, A., Dunnett, L., Fedorov, O., Ferla, M. P., Fuchs, M. R., Gorrie-Stone, T. J., Holton, J. M., Johnson, M. G., Krojer, T., Meigs, G., Powell, A. J., Rack, J. G. M., Rangel, V. L., Russi, S., Skyner, R. E., Smith, C. A., Soares, A. S., Wierman, J. L., Zhu, K., O’Brien, P., Jura, N., Ashworth, A., Irwin, J. J., Thompson, M. C., Gestwicki, J. E., von Delft, F., Shoichet, B. K., Fraser, J. S. & Ahel, I. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci Adv 7, (2021).
[3] Erlanson, D. A., McDowell, R. S. & O’Brien, T. Fragment-based drug discovery. J. Med. Chem. 47, 3463–3482 (2004).
[4] Murray, C. W. & Rees, D. C. The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192 (2009).
[5] Pearce, N. M., Krojer, T. & von Delft, F. Proper modelling of ligand binding requires an ensemble of bound and unbound states. Acta Crystallogr D Struct Biol 73, 256–266 (2017).
[6] Gahbauer, S., Correy, G. J., Schuller, M., Ferla, M. P., Doruk, Y. U., Rachman, M., Wu, T., Diolaiti, M., Wang, S., Jeffrey Neitz, R., Fearon, D., Radchenko, D., Moroz, Y., Irwin, J. J., Renslo, A. R., Taylor, J. C., Gestwicki, J. E., von Delft, F., Ashworth, A., Ahel, I., Shoichet, B. K. & Fraser, J. S. Structure-based inhibitor optimization for the Nsp3 Macrodomain of SARS-CoV-2. bioRxiv 2022.06.27.497816 (2022). doi:10.1101/2022.06.27.497816

